Example Data: Post-Diet Weights
Download example data UCLA website
Information about data:
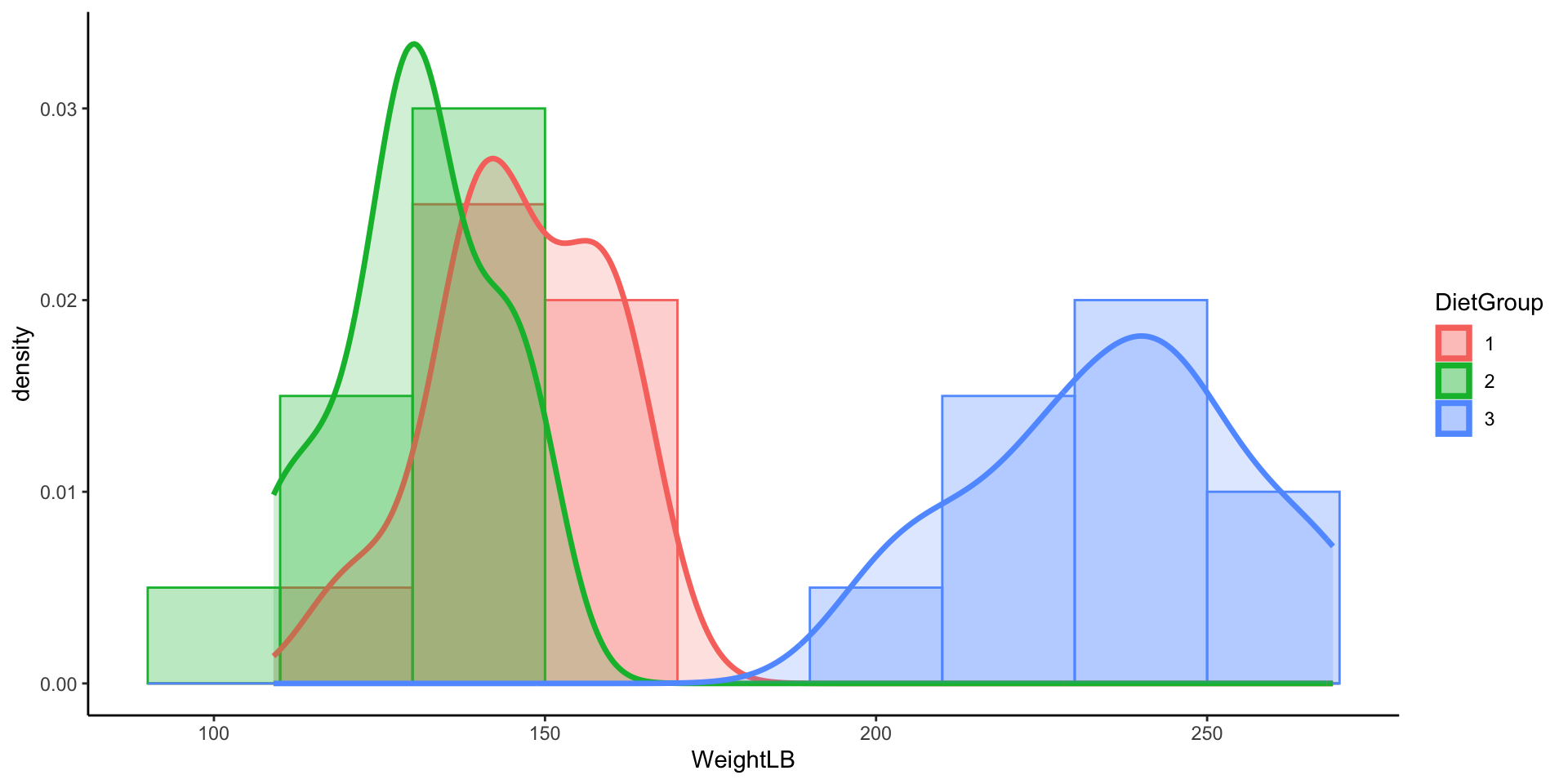
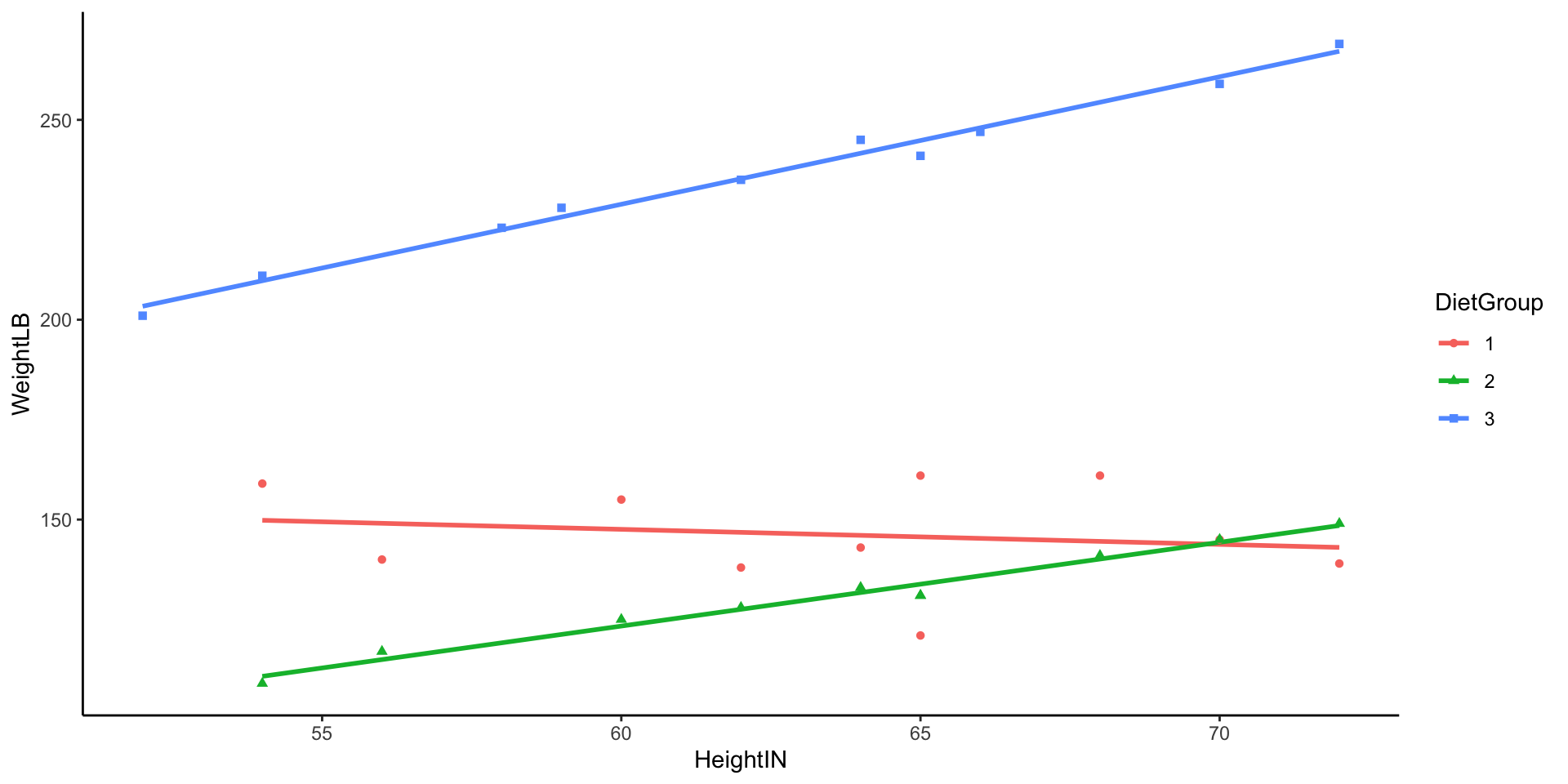
- In this example data file, it contains 30 subjects who used one of three diets: diet 1 (diet=1), diet 2 (diet=2), and a control group (diet=3).
- The file DietData.csv contains the data we needed.
- Variables in the data set are:
- Respondent: Respondent ID 1-30
- DietGroup: 1, 2, 3 representing the group to which a respondent was assigned
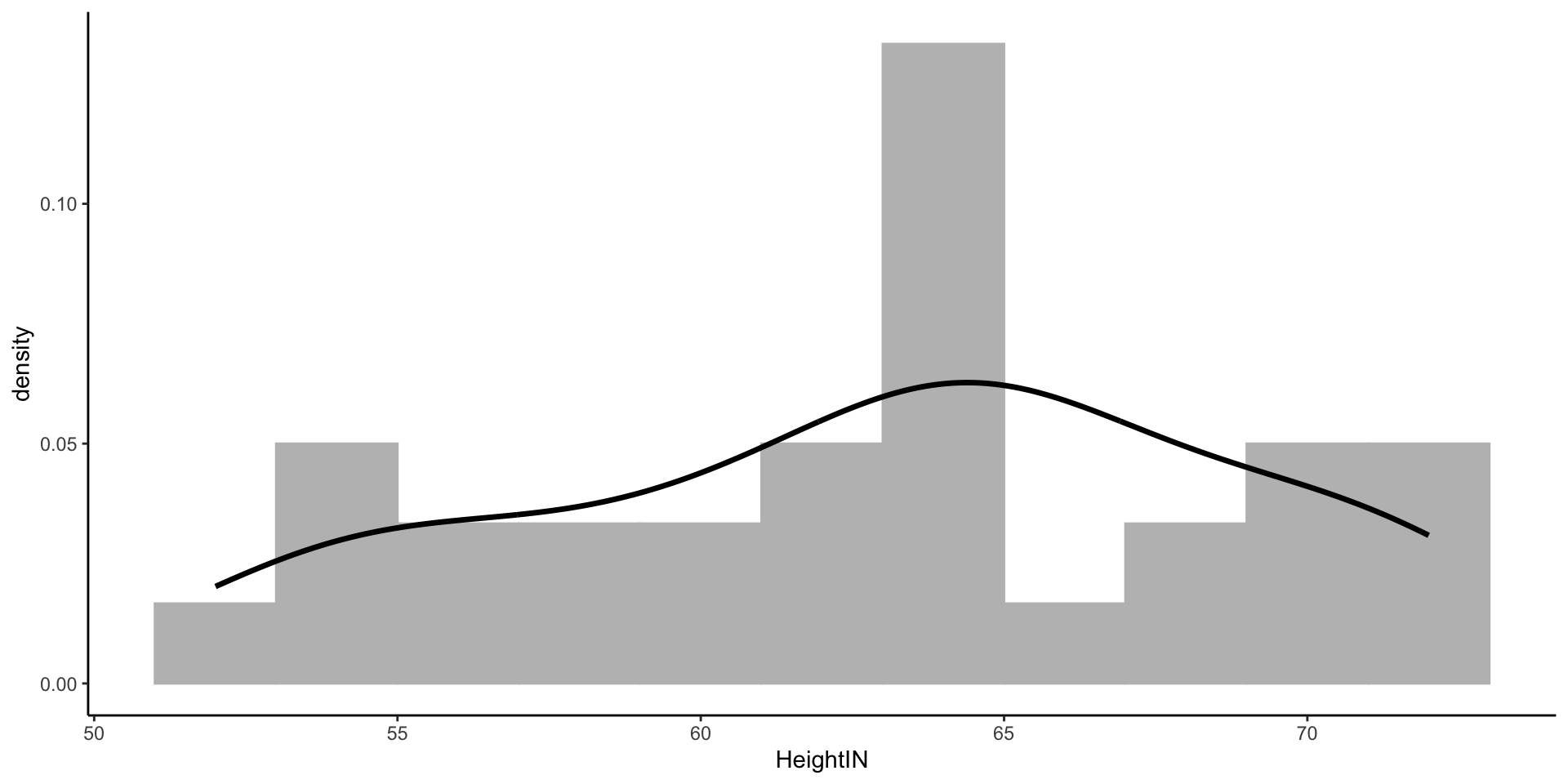
- HeightIN: The respondents’ height in inches
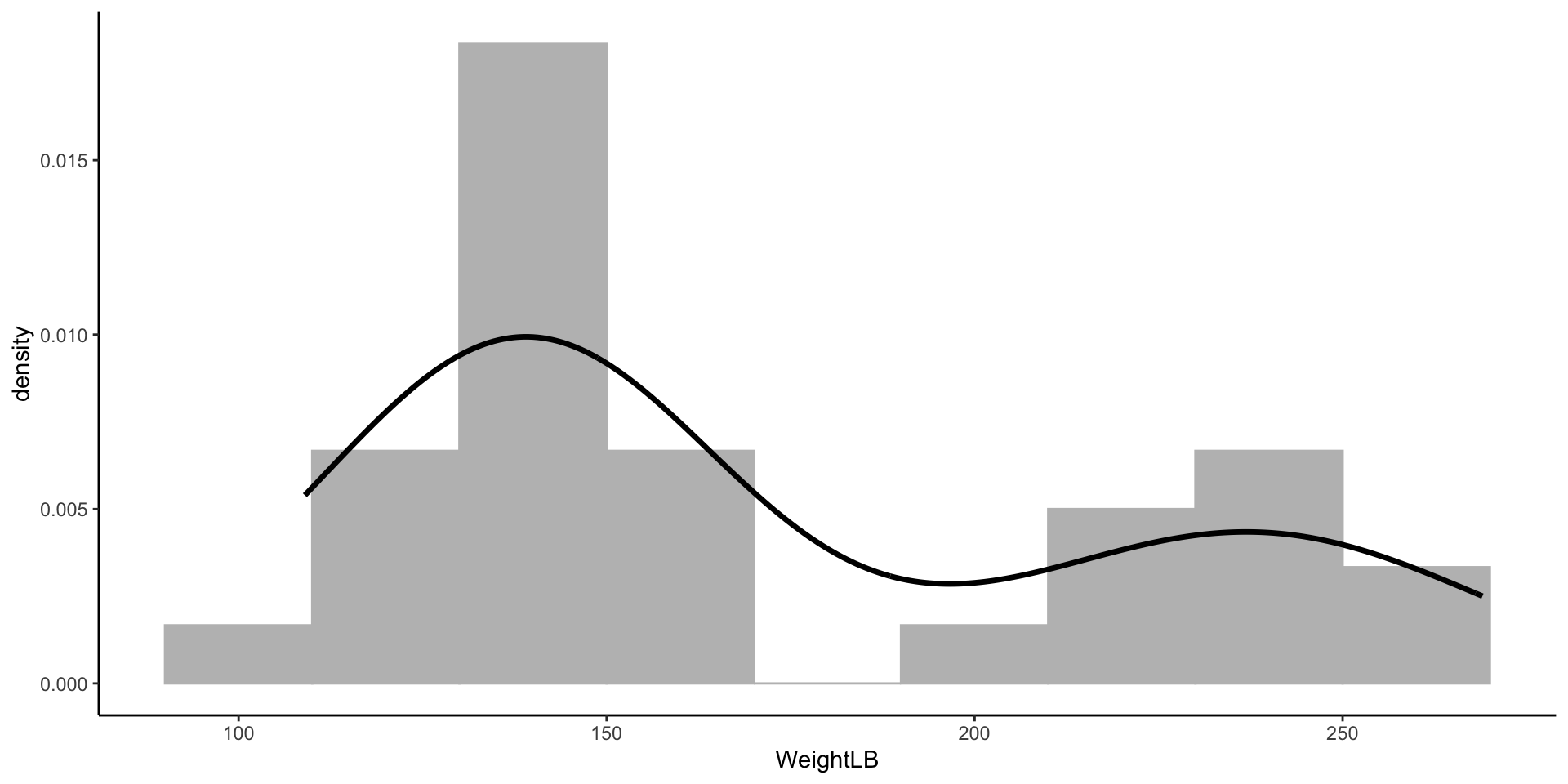
- WeightLB (Dependent Variable): The respondents’ weight in pounds
- Research Question: Are there differences in final weights between the three diet groups, and, if so, what the nature of the differences?
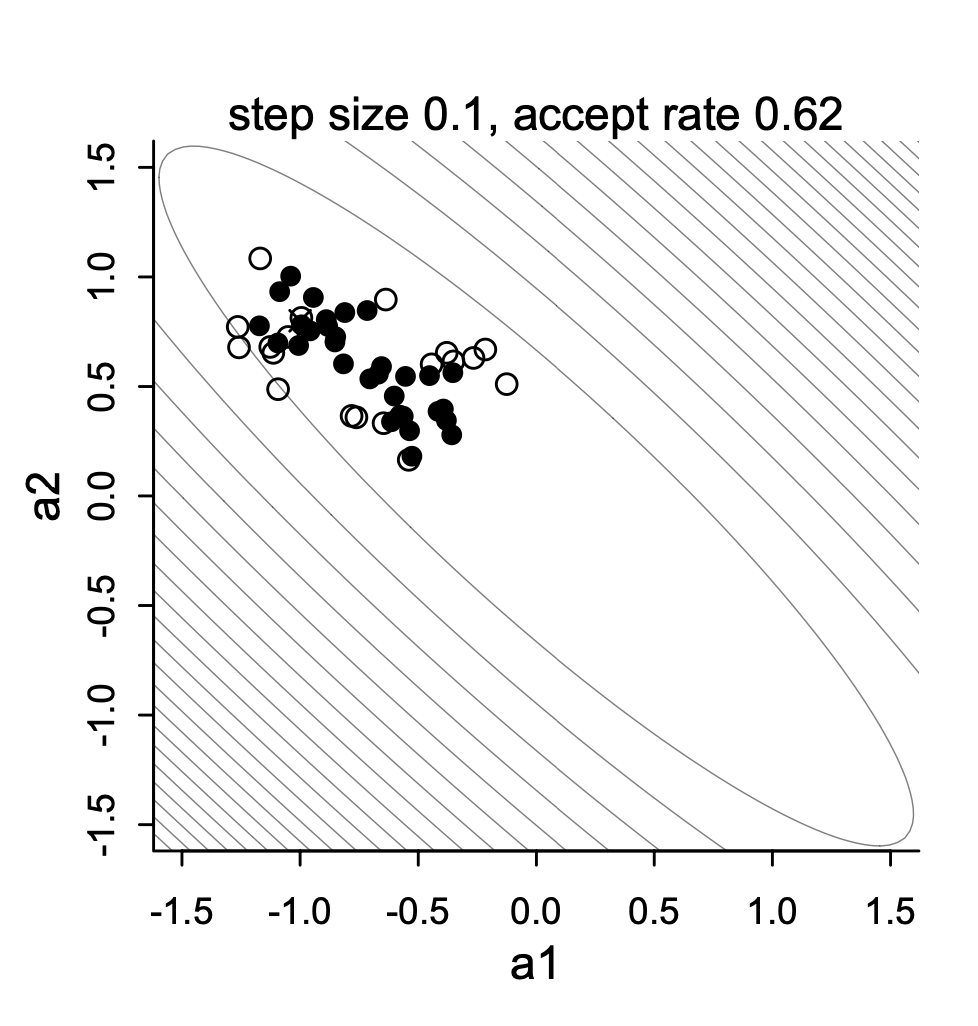
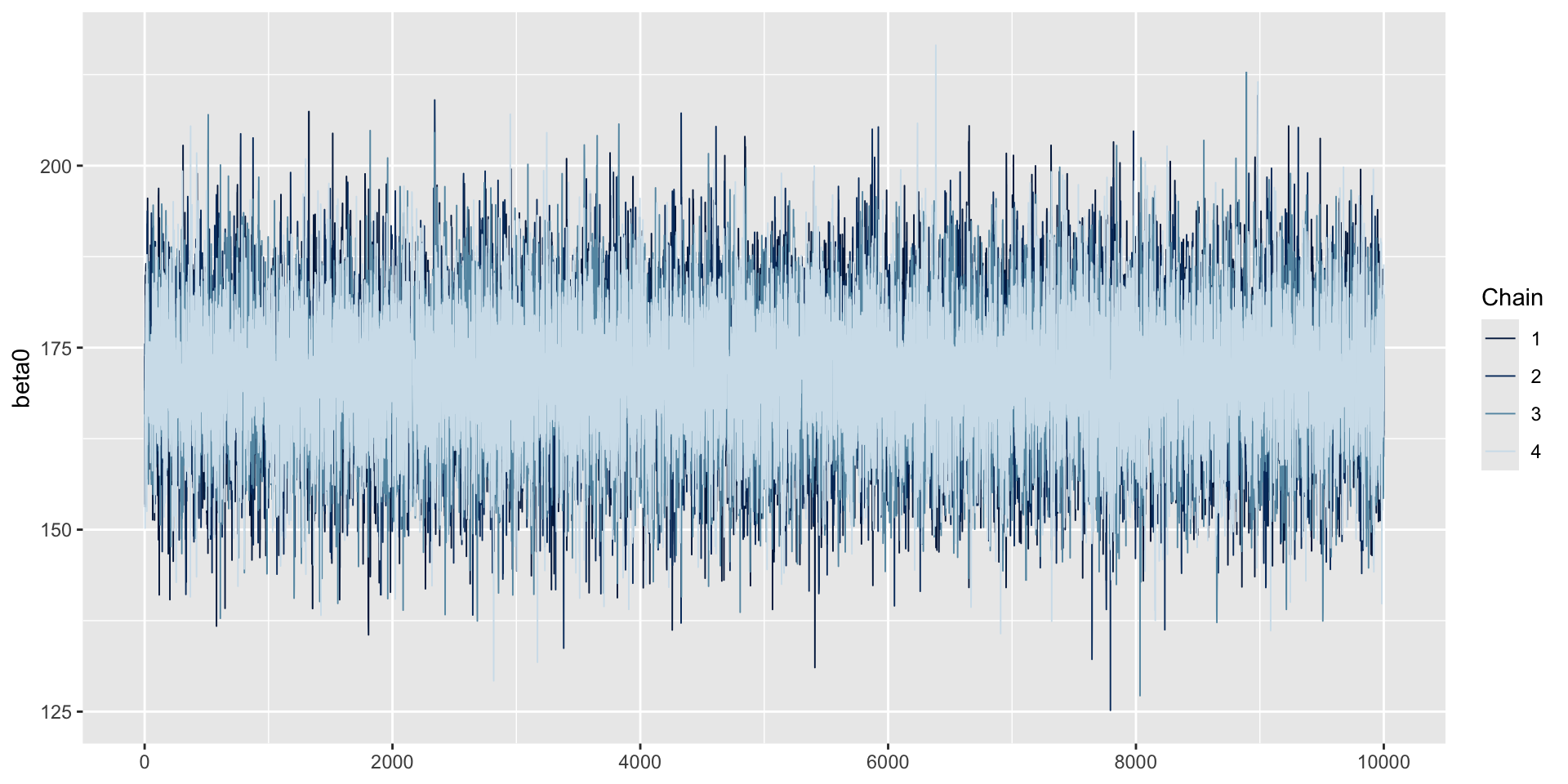
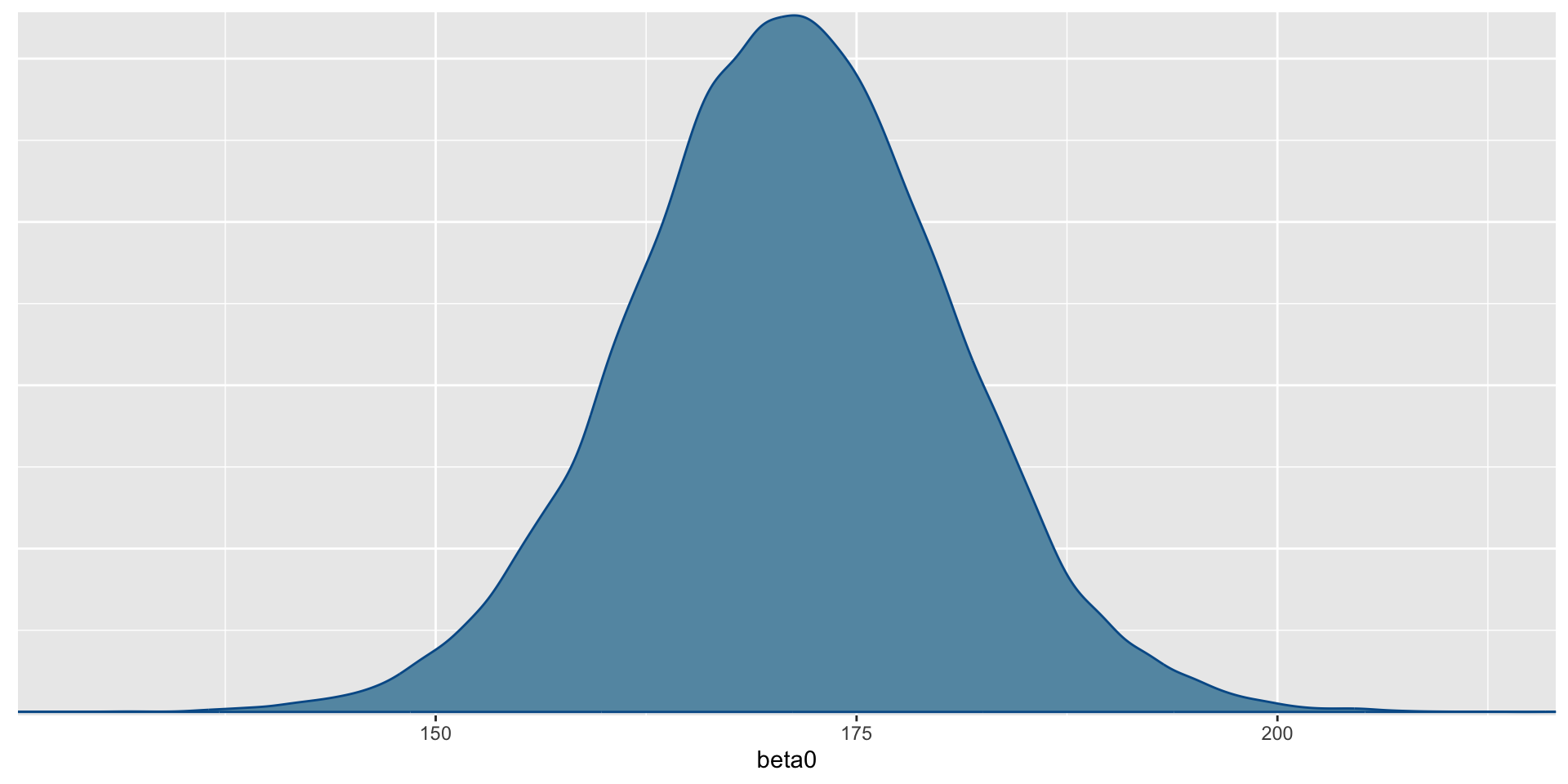
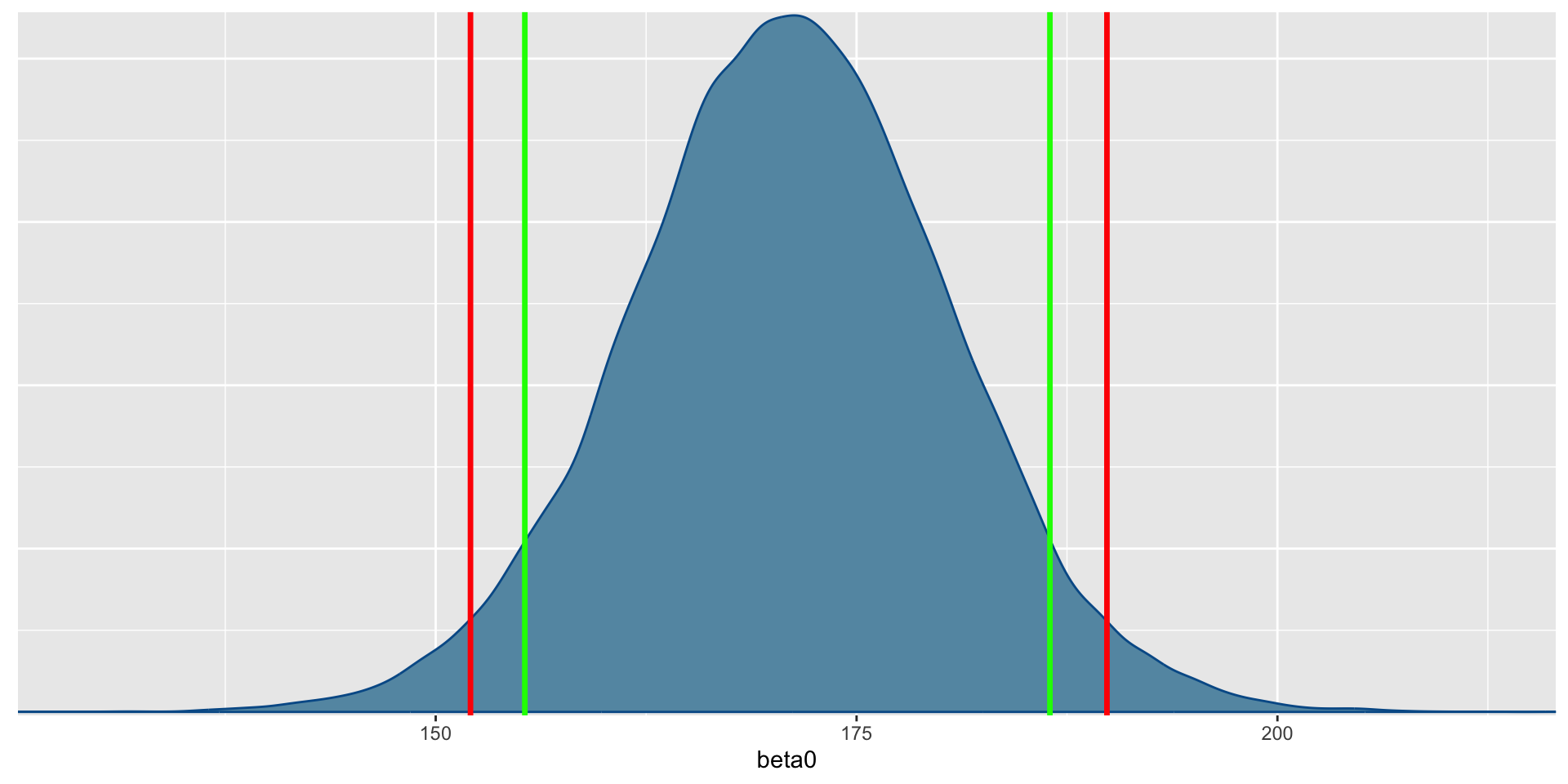
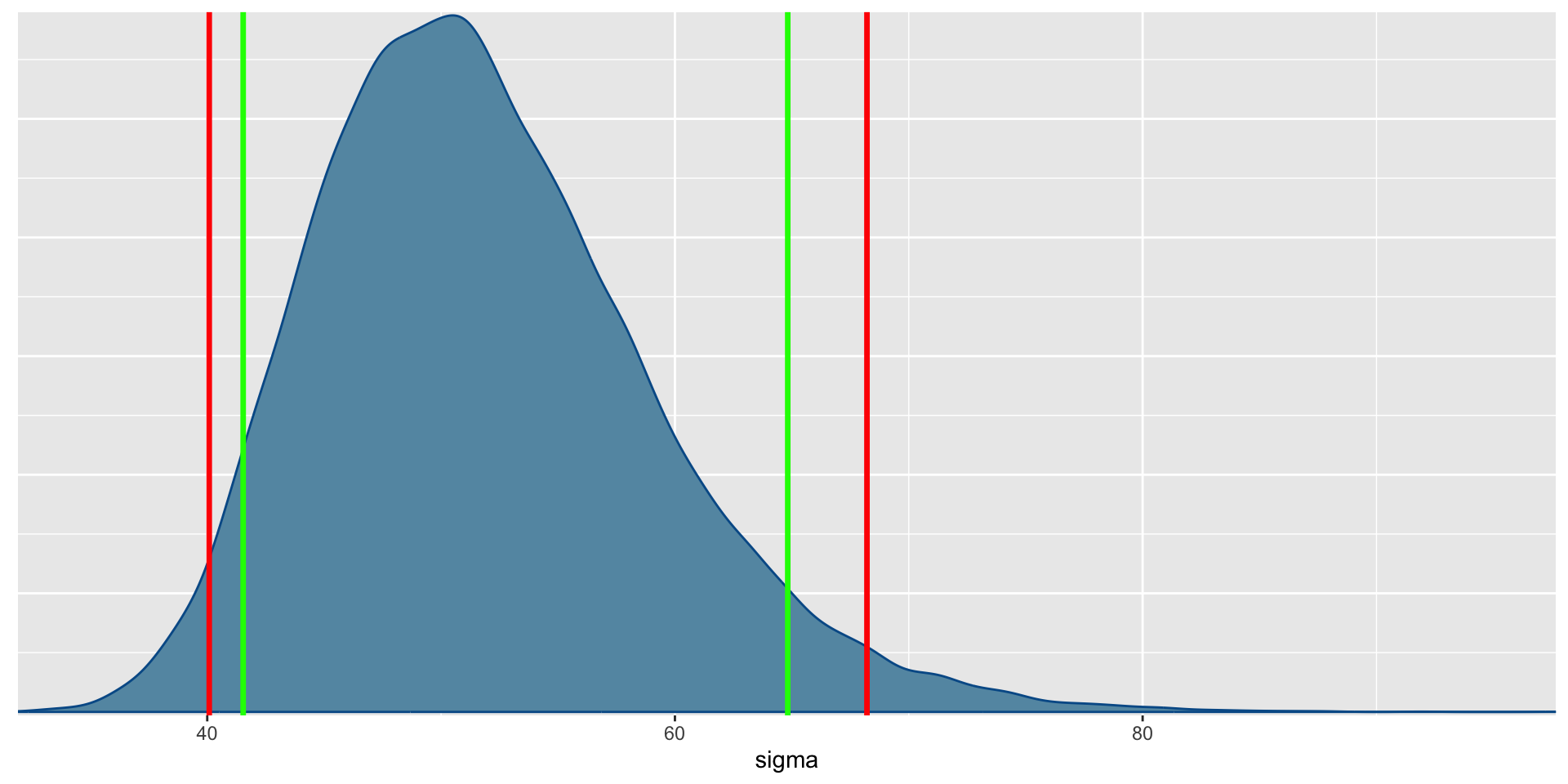
- Before we conduct the analysis, let’s look at the data