True Covariance Matrix - \(\bb{\Sigma}\)
[,1] [,2] [,3]
[1,] 1.00 -0.26 0.31
[2,] -0.26 1.00 -0.08
[3,] 0.31 -0.08 1.00Network Psychometrics
Educational Statistics and Research Methods
Understand what is network psychometrics and
Understand the relationship between psychometric/psychological network with factor analysis
Understand how network model can be applied into real scenarios
Network analysis is a broad area. It has many names in varied fields:
1 and 2 focus on the probabilistic relationships and further casual relationships among variables. 3 and 4 focuses on network structure and node importance. 5 focus on the regression coefficients of structural and measurement mdoel.
All 5 analysis methods have a network-shaped diagram. Graphical modeling is a more “general” term that can comprise of the other network models.
Factor analysis (common factor model) assumes the associations between observed features can be explained by one or more common factors.
Psychometric network, however, assumes the associations between observed features ARE the reason of the development of depression. Or “depression” is the network itself.
Pyschometric network is exploratory by nature. To obtain a meaningful network structure, psychometric networks need to drop weak edges but keep strong edges.
This procedure is typically called edge selection. One popular edge selection method is regularization.


Psychometric network analysis methodology includes steps of network structure estimation (to construct the network), network description (to characterize the network) and network stability analysis (to assess the robustness of results).
GGM is one type of Pairwise Markov random field (PMRF) when data are continuous.
In a PMRF, the joint likelihood of multivariate data is modelled through the use of pairwise conditional associations, leading to a network representation that is undirected.
For \(p\)-dimensional data following multivariate normal distribution:
\[ \newcommand{\bb}[1]{\boldsymbol{#1}} \bb{X} \sim \mathcal{N}(\mu, \bb{K}^{-1}) \]
Where \(K\) is a inverse covariance matrix of \(\bb{X}\) (\(K = \Sigma^{-1}\)), also known as precision/concentration matrix.
To obtain sparse network structure, the \(i\)th row and \(j\)th column element of \(\bb{K}\), \(k_{ij}=0\) when edge \(\{j, k\}\) is not included in the network \(G\),
GGM can be standardized as the partial correlation network, in which each edge of GGM representing partial correlations between two nodes.
\[ \rho_{ij}=\rho(X^{(i)}, X^{(j)}|\bb{X}^{-(i,j)}) = -\frac{k_{ij}}{\sqrt{k_{ii}}\sqrt{k_{jj}}} \]
Assume there are three variables: fatigue, insomnia, concentration
[,1] [,2] [,3]
[1,] 1.00 -0.26 0.31
[2,] -0.26 1.00 -0.08
[3,] 0.31 -0.08 1.00Factor analysis model:
\[ \bb{X}\sim\mathcal{N}(\mu, \bb{\Lambda\Psi\Lambda^\text{T}+\Phi}) \]
GGM with partial correlation matrix:
\[ \bb{X} \sim \mathcal{N}(0, \bb{\Delta(I-\Omega)^{-1}\Delta}) \]
Where
psychonetrics: Partial correlation matrix estimation [,1] [,2] [,3]
[1,] 1.00 -0.26 0.31
[2,] -0.26 1.00 -0.08
[3,] 0.31 -0.08 1.00 [,1] [,2] [,3]
[1,] 0.000 -0.248 0.300
[2,] -0.248 0.000 0.001
[3,] 0.300 0.001 0.000 [,1] [,2] [,3]
[1,] 0.921 0.000 0.000
[2,] 0.000 0.966 0.000
[3,] 0.000 0.000 0.951BGGM: Bayesian approach [,1] [,2] [,3]
[1,] 0.000 -0.258 0.189
[2,] -0.258 0.000 0.040
[3,] 0.189 0.040 0.000BGGM: Bayesian Gaussian Graphical Models
---
Type: continuous
Analytic: FALSE
Formula:
Posterior Samples: 1000
Observations (n):
Nodes (p): 3
Relations: 3
---
Call:
BGGM::estimate(Y = dat, type = "continuous", analytic = FALSE,
iter = 1000)
---
Estimates:
Relation Post.mean Post.sd Cred.lb Cred.ub
1--2 -0.258 0.042 -0.342 -0.173
1--3 0.189 0.043 0.105 0.274
2--3 0.040 0.046 -0.055 0.129
--- Multiple procedure and software can be used to perform edge selection:
prune function in psychonetrics package uses stepdown model search by pruning non-significant parameters
EBICglasso in qgraph and glasso package uses Extended Bayesian Information Criterion (EBIC) to select best model
\[ \text{EBIC}=-2\text{L}+E(\log(N))+4\gamma E(log(P)) \]
select function in BGGM package uses Bayesian Hypothesis Testing — Bayes Factor (BF) to select model
25 self-reported personality items representing 5 factors.
70.6% edges are nonzero using EBICglasso, while only 63.6% edges are nonzero using BF method and 62.0% edges are kept using \(\alpha =.01\) significance testing
library(psych)
library(qgraph)
data(bfi)
big5groups <- list(
Agreeableness = 1:5,
Conscientiousness = 6:10,
Extraversion = 11:15,
Neuroticism = 16:20,
Openness = 21:25
)
CorMat <- cor_auto(bfi[,1:25])
EBICgraph <- EBICglasso(CorMat, nrow(bfi), 0.5, threshold = TRUE)
## density
density_nonzero_edge <- function(pcor_matrix){
N_nonzero_edge = (sum(pcor_matrix == 0) - ncol(pcor_matrix)) /2
N_all_edge = ncol(pcor_matrix)*(ncol(pcor_matrix)-1)/2
N_nonzero_edge/N_all_edge
}
density_nonzero_edge(EBICgraph)[1] 0.7066667[1] 0.62


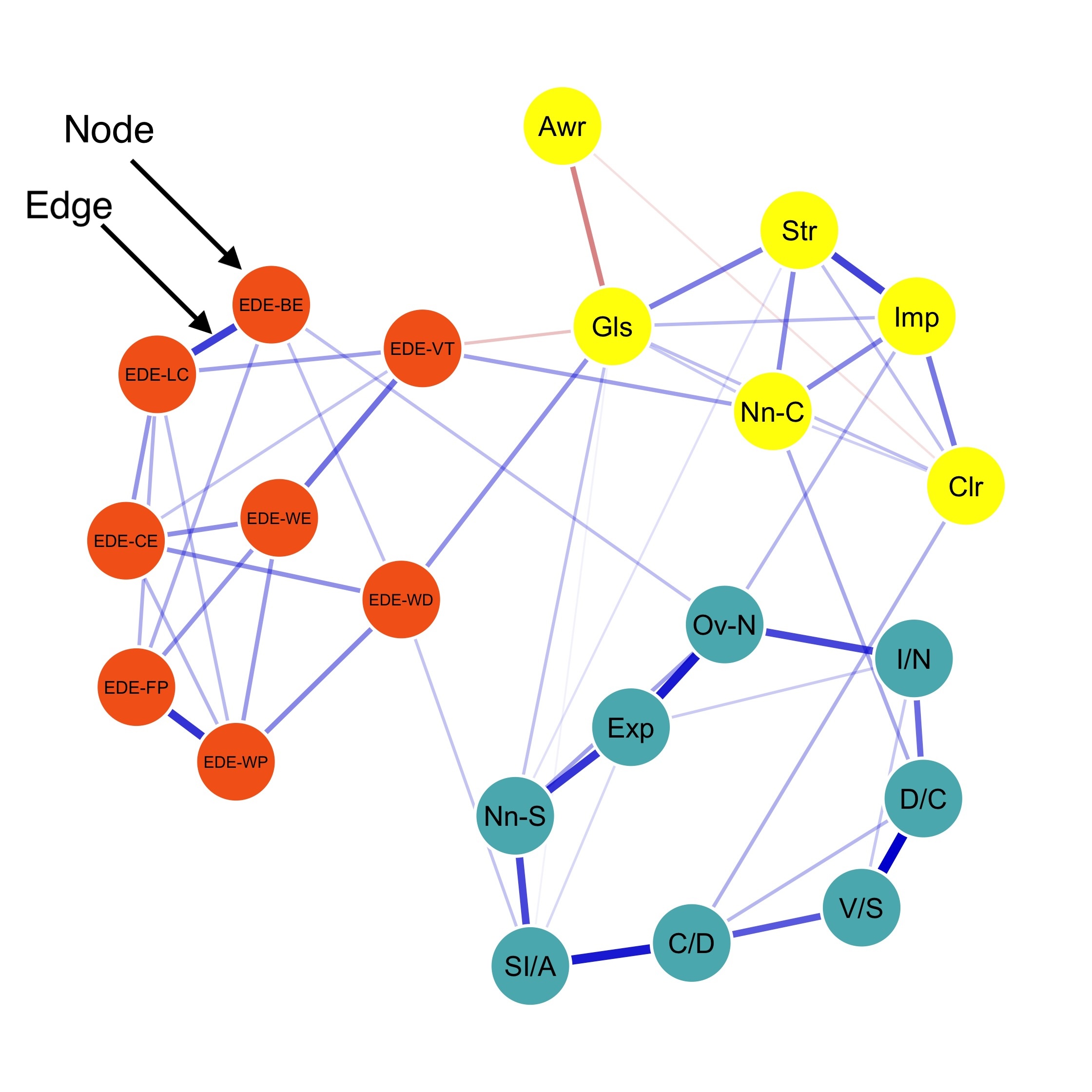
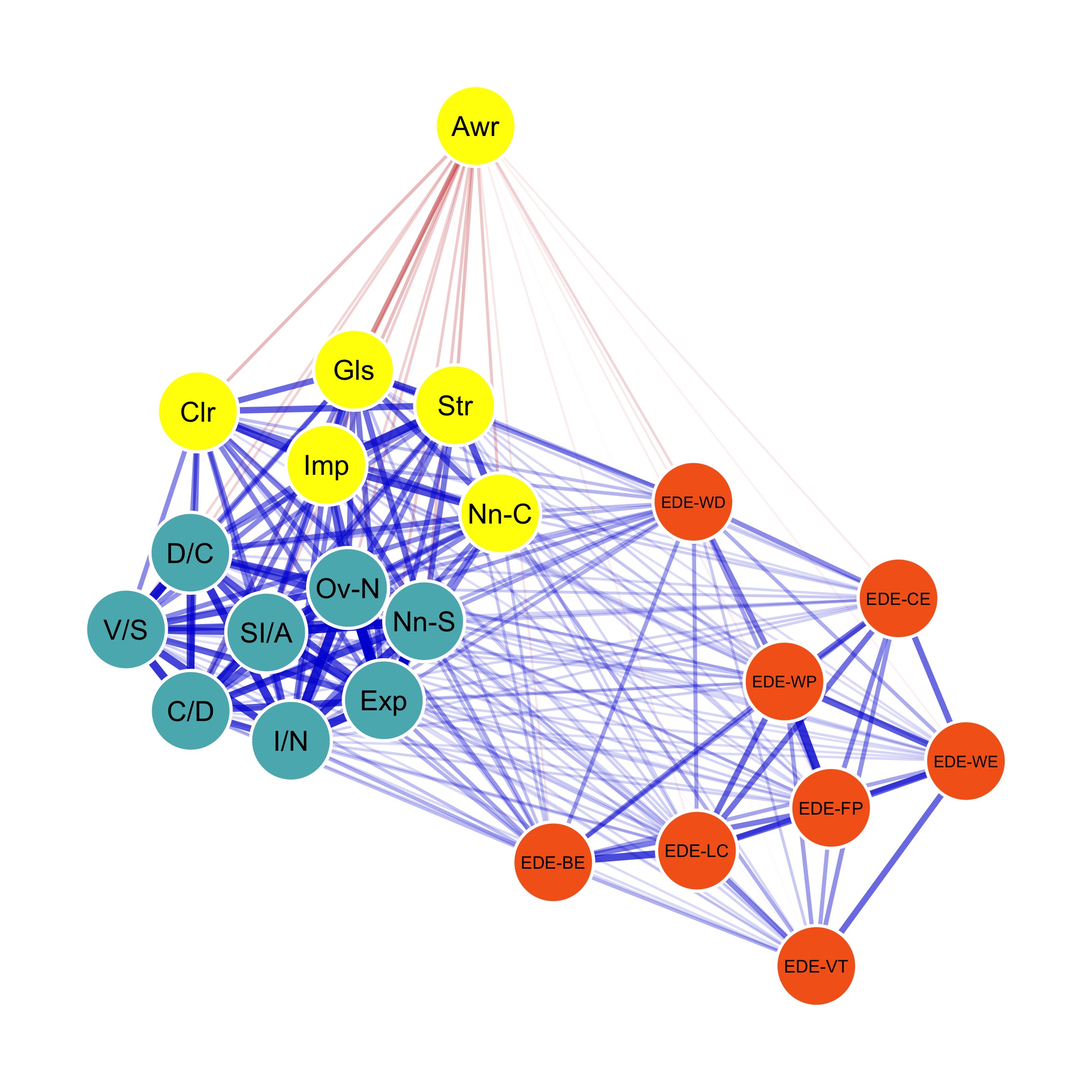
Strength centrality measures suggest that C4 and E4 or N1 have highest centrality indicating they play most imporatant roles in the networks.



E4 and E5 has highest bridge strength, indicating they serve as bridges linking communities of personality. They are important elements connecting varied types of personality


