Lecture 07
Generalized Measurement Models: Modeling Observed Data
Generalized Measurement Models: Modeling Observed Data
Educational Statistics and Research Methods
Today’s example is from a bootstrap resample of 177 undergraduate students at a large state university in the Midwest.
The survey was a measure of 10 questions about their beliefs in various conspiracy theories that were being passed around the internet in the early 2010s
All item responses were on a 5-point Likert scale with:
Strong Disagree
Disagree
Neither Agree nor Disagree
Agree
Strongly Agree
The purpose of this survey was to study individual beliefs regarding conspiracies.
Our purpose in using this instrument is to provide a context that we all may find relevant as many of these conspiracies are still prevalent.
Billionaire George Soros is behind a hidden plot to destabilize the American government, take control of the media, and put the world under his control.
The U.S. government is mandating the switch to compact fluorescent light bulbs because such lights make people more obedient and easier to control.
Government officials are covertly Building a 12-lane “NAFTA superhighway” that runs from Mexico to Canada through America’s heartland.
Government officials purposely developed and spread drugs like crack-cocaine and diseases like AIDS in order to destroy the African American community.
God sent Hurricane Katrina to punish America for its sins.
All items seem to be positive skewed
## ── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
## ✔ dplyr 1.1.4 ✔ readr 2.1.5
## ✔ forcats 1.0.0 ✔ stringr 1.5.1
## ✔ ggplot2 3.5.1 ✔ tibble 3.2.1
## ✔ lubridate 1.9.4 ✔ tidyr 1.3.1
## ✔ purrr 1.0.4
## ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
## ✖ dplyr::filter() masks stats::filter()
## ✖ dplyr::lag() masks stats::lag()
## ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors
## here() starts at /Users/jihong/Documents/Projects/website-jihongFor today’s lecture, we will assume each of the 10 items measures one single latent variable
θ: tendency to believe in conspiracy theories
Higher value of θ suggests more likelihood of believing in conspiracy theories
Let’s denote this latent variable as θp for individual p
We will assume this latent variable is:
Continuous
Normally distribution: θp∼N(μθ,σθ)
Across all people, we will denote the set of vector of latent variable as
Θ=[θ1,⋯,θP]T
A psychometric model posits that one or more hypothesized latent variable(s) is the common cause that can predict a person’s response to observed items:
UnidimensionalA typical linear regression is like
Yp=β0+β1Xp+ep
with ep∼N(0,σe)
If we replace Xp with latent variable θp, and replace β as factor loading λ
We can get the linear regression function (IRF) for each item
Yp1=μ0+λ1θp+ep1; ep1∼N(0,ψ21)Yp2=μ2+λ2θp+ep2; ep2∼N(0,ψ22)Yp3=μ3+λ3θp+ep3; ep3∼N(0,ψ23)Yp4=μ4+λ4θp+ep4; ep4∼N(0,ψ24)Yp5=μ5+λ5θp+ep5; ep5∼N(0,ψ25)Yp6=μ6+λ6θp+ep6; ep6∼N(0,ψ26)Yp7=μ7+λ7θp+ep7; ep7∼N(0,ψ27)Yp8=μ8+λ8θp+ep8; ep8∼N(0,ψ28)Yp9=μ9+λ9θp+ep9; ep9∼N(0,ψ29)Yp10=μ10+λ10θp+ep10; ep10∼N(0,ψ102)
μi: Item intercept
Interpretation: the expected score on the item i when θp=0
Higher Item intercept suggests more likely to believe in conspiracy for people with average level of conspiracy belief
So it is also called item easiness in item response theory (IRT)
λi: Factor loading or Item discrimination
ψ2i: Unique variance1
When we specify measurement model, we need to choose on scale identification method for latent variable
In this study, we assume θp∼N(0,1) which allows us to estimate all item parameters of the model
This is what we call a standardization identification method
Factor scores are like Z-scores
Recall that we can use matrix operation to make Stan estimate psychometric models with normal outcomes:
The model (predictor) matrix cannot be used
The data will be imported as a matrix
The parameters will be specified as vectors of each type
Each item will have its own set of parameters
Implications for the use of prior distributions
data Blockdata {
1 int<lower=0> nObs; // number of observations
2 int<lower=0> nItems; // number of items
matrix[nObs, nItems] Y; // item responses in a matrix
vector[nItems] meanMu;
3 matrix[nItems, nItems] covMu; // prior covariance matrix for coefficients
vector[nItems] meanLambda; // prior mean vector for coefficients
4 matrix[nItems, nItems] covLambda; // prior covariance matrix for coefficients
5 vector[nItems] psiRate; // prior rate parameter for unique standard deviations
}nObs is 177, declared as integer with lower bound as 0
nItems is 11, declared as integer with lower bound as 0
meanMu as covMu are prior mean and covariance matrix for μi
meanLambda and covLambda are prior mean and covariance matrix for λi
psiRate is prior rate parameter for ψi
parameter Blockparameters {
vector[nObs] theta; // the latent variables (one for each person)
vector[nItems] mu; // the item intercepts (one for each item)
vector[nItems] lambda; // the factor loadings/item discriminations (one for each item)
vector<lower=0>[nItems] psi; // the unique standard deviations (one for each item)
}Here, the parameterization of λ (factor loadings / item discrimination) can lead to problems in estimation
The issue: λiθp=(−λi)(−θp)
To demonstrate, we will start with different random number seed
Currently using 09102022: works fine
Change to 25102022: big problem
model {
lambda ~ multi_normal(meanLambda, covLambda); // Prior for item discrimination/factor loadings
mu ~ multi_normal(meanMu, covMu); // Prior for item intercepts
psi ~ exponential(psiRate); // Prior for unique standard deviations
theta ~ normal(0, 1); // Prior for latent variable (with mean/sd specified)
for (item in 1:nItems){
Y[,item] ~ normal(mu[item] + lambda[item]*theta, psi[item]);
}
}The loop here conducts the model via item response function (IRF) for each item:
Assumption of conditional independence enables this
The item mean is set by the conditional mean of the model
The loop puts each item’s parameters into the question
There is not uniform agreement about the choices of prior distributions for item parameters
We will use uninformative priors on each to begin
For now:
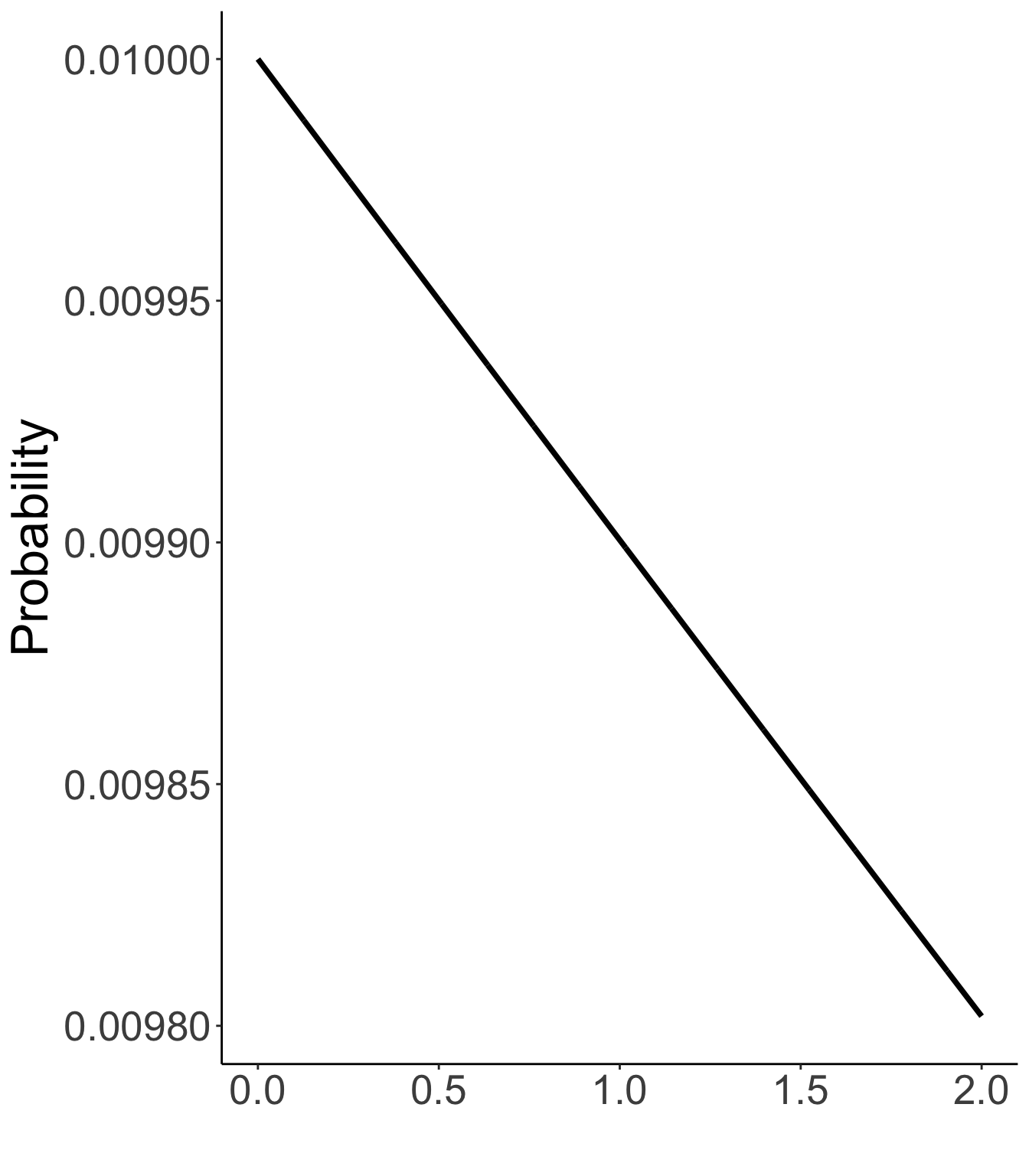
Item intercepts: μi∼N(0,σ2μi=1000)
Factor loadings / item discrimination: λi∼N(0,σ2λi=1000)
Unique standard deviations: ψi∼exponential(rψi=.01)
Prior distribution for item intercepts, factor loadings
N(0, 1000)
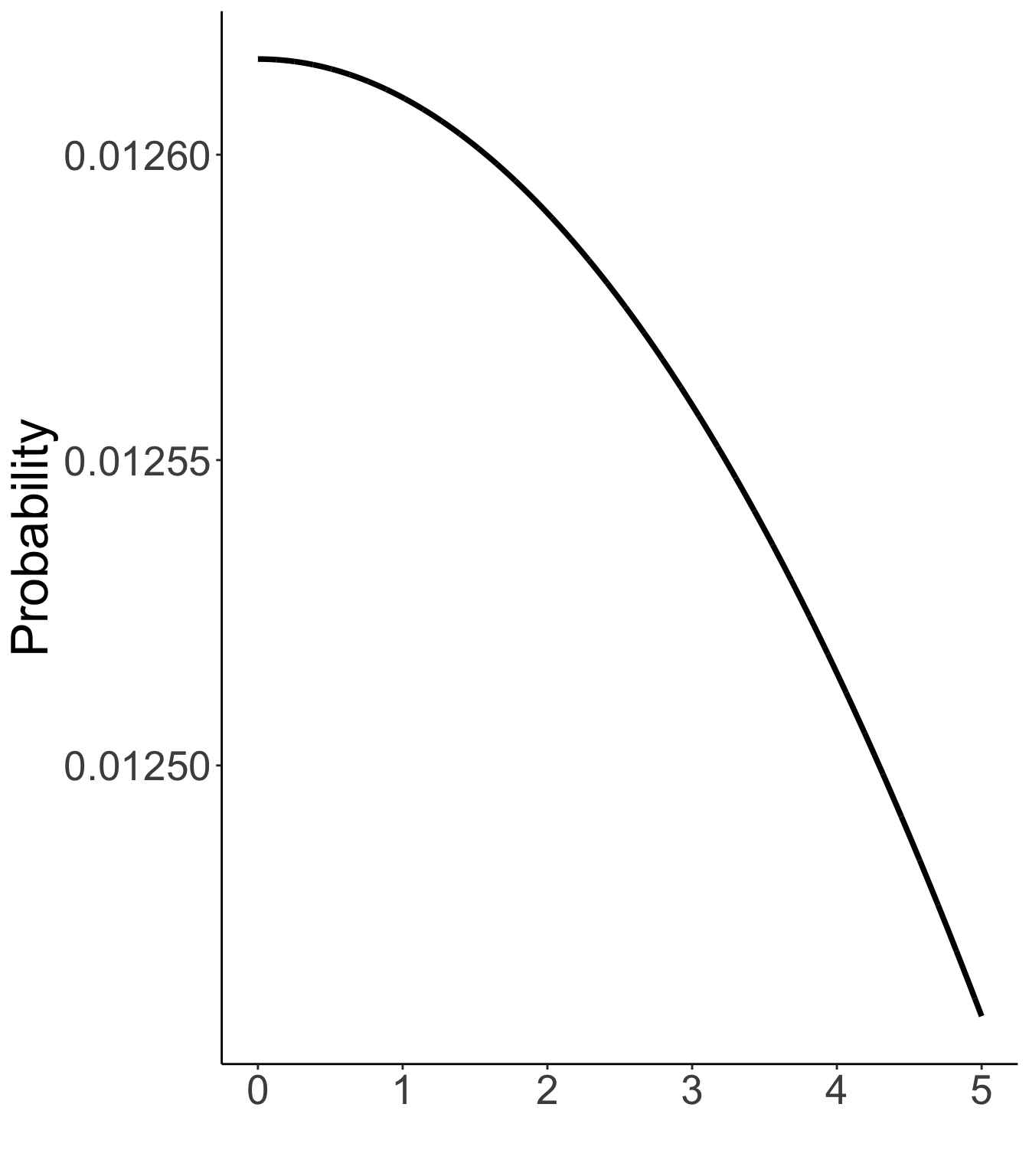
Prior distribution of unique standard deviations
Exp(.01)
nObs = nrow(conspiracyItems)
nItems = ncol(conspiracyItems)
# item intercept hyperparameters
muMeanHyperParameter = 0
muMeanVecHP = rep(muMeanHyperParameter, nItems)
muVarianceHyperParameter = 1000
muCovarianceMatrixHP = diag(x = sqrt(muVarianceHyperParameter), nrow = nItems)
# item discrimination/factor loading hyperparameters
lambdaMeanHyperParameter = 0
lambdaMeanVecHP = rep(lambdaMeanHyperParameter, nItems)
lambdaVarianceHyperParameter = 1000
lambdaCovarianceMatrixHP = diag(x = sqrt(lambdaVarianceHyperParameter), nrow = nItems)
# unique standard deviation hyperparameters
psiRateHyperParameter = .01
psiRateVecHP = rep(.1, nItems)
modelCFA_data = list(
nObs = nObs,
nItems = nItems,
Y = conspiracyItems,
meanMu = muMeanVecHP,
covMu = muCovarianceMatrixHP,
meanLambda = lambdaMeanVecHP,
covLambda = lambdaCovarianceMatrixHP,
psiRate = psiRateVecHP
)Note: In Stan, the second argument to the “normal” function is the standard deviation (i.e., the scale), not the variance (as in Bayesian Data Analysis) and not the inverse-variance (i.e., precision) (as in BUGS).
The total number of parameters is 207.
[1] "lp__" "theta[1]" "theta[2]" "theta[3]" "theta[4]"
[6] "theta[5]" "theta[6]" "theta[7]" "theta[8]" "theta[9]"
[11] "theta[10]" "theta[11]" "theta[12]" "theta[13]" "theta[14]"
[16] "theta[15]" "theta[16]" "theta[17]" "theta[18]" "theta[19]"
[21] "theta[20]" "theta[21]" "theta[22]" "theta[23]" "theta[24]"
[26] "theta[25]" "theta[26]" "theta[27]" "theta[28]" "theta[29]"
[31] "theta[30]" "theta[31]" "theta[32]" "theta[33]" "theta[34]"
[36] "theta[35]" "theta[36]" "theta[37]" "theta[38]" "theta[39]"
[41] "theta[40]" "theta[41]" "theta[42]" "theta[43]" "theta[44]"
[46] "theta[45]" "theta[46]" "theta[47]" "theta[48]" "theta[49]"
[51] "theta[50]" "theta[51]" "theta[52]" "theta[53]" "theta[54]"
[56] "theta[55]" "theta[56]" "theta[57]" "theta[58]" "theta[59]"
[61] "theta[60]" "theta[61]" "theta[62]" "theta[63]" "theta[64]"
[66] "theta[65]" "theta[66]" "theta[67]" "theta[68]" "theta[69]"
[71] "theta[70]" "theta[71]" "theta[72]" "theta[73]" "theta[74]"
[76] "theta[75]" "theta[76]" "theta[77]" "theta[78]" "theta[79]"
[81] "theta[80]" "theta[81]" "theta[82]" "theta[83]" "theta[84]"
[86] "theta[85]" "theta[86]" "theta[87]" "theta[88]" "theta[89]"
[91] "theta[90]" "theta[91]" "theta[92]" "theta[93]" "theta[94]"
[96] "theta[95]" "theta[96]" "theta[97]" "theta[98]" "theta[99]"
[101] "theta[100]" "theta[101]" "theta[102]" "theta[103]" "theta[104]"
[106] "theta[105]" "theta[106]" "theta[107]" "theta[108]" "theta[109]"
[111] "theta[110]" "theta[111]" "theta[112]" "theta[113]" "theta[114]"
[116] "theta[115]" "theta[116]" "theta[117]" "theta[118]" "theta[119]"
[121] "theta[120]" "theta[121]" "theta[122]" "theta[123]" "theta[124]"
[126] "theta[125]" "theta[126]" "theta[127]" "theta[128]" "theta[129]"
[131] "theta[130]" "theta[131]" "theta[132]" "theta[133]" "theta[134]"
[136] "theta[135]" "theta[136]" "theta[137]" "theta[138]" "theta[139]"
[141] "theta[140]" "theta[141]" "theta[142]" "theta[143]" "theta[144]"
[146] "theta[145]" "theta[146]" "theta[147]" "theta[148]" "theta[149]"
[151] "theta[150]" "theta[151]" "theta[152]" "theta[153]" "theta[154]"
[156] "theta[155]" "theta[156]" "theta[157]" "theta[158]" "theta[159]"
[161] "theta[160]" "theta[161]" "theta[162]" "theta[163]" "theta[164]"
[166] "theta[165]" "theta[166]" "theta[167]" "theta[168]" "theta[169]"
[171] "theta[170]" "theta[171]" "theta[172]" "theta[173]" "theta[174]"
[176] "theta[175]" "theta[176]" "theta[177]" "mu[1]" "mu[2]"
[181] "mu[3]" "mu[4]" "mu[5]" "mu[6]" "mu[7]"
[186] "mu[8]" "mu[9]" "mu[10]" "lambda[1]" "lambda[2]"
[191] "lambda[3]" "lambda[4]" "lambda[5]" "lambda[6]" "lambda[7]"
[196] "lambda[8]" "lambda[9]" "lambda[10]" "psi[1]" "psi[2]"
[201] "psi[3]" "psi[4]" "psi[5]" "psi[6]" "psi[7]"
[206] "psi[8]" "psi[9]" "psi[10]" Different seed will have different initial values that may leads to convergence.
cmdstanr sampling call:$total
[1] 2.050416
$chains
chain_id warmup sampling total
1 1 0.663 1.269 1.932
2 2 0.675 1.278 1.953
3 3 0.668 1.264 1.932
4 4 0.671 1.269 1.940Checking convergence with ˆR (PSRF):
[1] 1.007473
Attaching package: 'kableExtra'The following object is masked from 'package:dplyr':
group_rows| variable | mean | median | sd | mad | q5 | q95 | rhat | ess_bulk | ess_tail |
|---|---|---|---|---|---|---|---|---|---|
| mu[1] | 2.36 | 2.36 | 0.09 | 0.09 | 2.22 | 2.50 | 1.00 | 1113.56 | 3020.86 |
| mu[2] | 1.95 | 1.95 | 0.08 | 0.08 | 1.81 | 2.09 | 1.01 | 687.12 | 2198.78 |
| mu[3] | 1.87 | 1.87 | 0.08 | 0.08 | 1.73 | 2.01 | 1.00 | 882.90 | 2490.04 |
| mu[4] | 2.01 | 2.01 | 0.08 | 0.08 | 1.87 | 2.15 | 1.00 | 782.88 | 2047.69 |
| mu[5] | 1.98 | 1.98 | 0.08 | 0.08 | 1.84 | 2.11 | 1.01 | 552.12 | 1622.83 |
| mu[6] | 1.89 | 1.89 | 0.08 | 0.08 | 1.76 | 2.01 | 1.01 | 599.94 | 1806.95 |
| mu[7] | 1.72 | 1.72 | 0.08 | 0.08 | 1.59 | 1.85 | 1.01 | 742.75 | 1914.12 |
| mu[8] | 1.84 | 1.84 | 0.07 | 0.07 | 1.72 | 1.96 | 1.01 | 585.20 | 1632.31 |
| mu[9] | 1.80 | 1.80 | 0.09 | 0.09 | 1.66 | 1.95 | 1.01 | 760.88 | 2237.89 |
| mu[10] | 1.52 | 1.52 | 0.08 | 0.08 | 1.39 | 1.65 | 1.00 | 1040.83 | 3151.00 |
| lambda[1] | 0.74 | 0.74 | 0.08 | 0.08 | 0.61 | 0.88 | 1.00 | 3088.26 | 3745.45 |
| lambda[2] | 0.87 | 0.87 | 0.08 | 0.08 | 0.75 | 1.00 | 1.00 | 2195.48 | 3294.83 |
| lambda[3] | 0.80 | 0.80 | 0.08 | 0.08 | 0.68 | 0.93 | 1.00 | 2378.66 | 4075.61 |
| lambda[4] | 0.84 | 0.84 | 0.08 | 0.08 | 0.72 | 0.97 | 1.00 | 2227.71 | 3241.24 |
| lambda[5] | 1.00 | 0.99 | 0.07 | 0.07 | 0.88 | 1.12 | 1.00 | 1428.73 | 2659.75 |
| lambda[6] | 0.90 | 0.90 | 0.06 | 0.06 | 0.80 | 1.01 | 1.00 | 1480.39 | 2228.33 |
| lambda[7] | 0.76 | 0.76 | 0.07 | 0.07 | 0.65 | 0.88 | 1.00 | 2106.97 | 3177.05 |
| lambda[8] | 0.85 | 0.85 | 0.06 | 0.06 | 0.76 | 0.95 | 1.00 | 1440.99 | 2422.00 |
| lambda[9] | 0.86 | 0.86 | 0.08 | 0.08 | 0.74 | 0.99 | 1.00 | 2228.18 | 3689.68 |
| lambda[10] | 0.67 | 0.67 | 0.08 | 0.08 | 0.55 | 0.80 | 1.00 | 3198.84 | 4681.76 |
| psi[1] | 0.89 | 0.89 | 0.05 | 0.05 | 0.81 | 0.98 | 1.00 | 15209.82 | 6290.16 |
| psi[2] | 0.73 | 0.73 | 0.04 | 0.04 | 0.67 | 0.81 | 1.00 | 12079.22 | 6272.07 |
| psi[3] | 0.78 | 0.78 | 0.05 | 0.04 | 0.71 | 0.86 | 1.00 | 13042.72 | 5945.80 |
| psi[4] | 0.76 | 0.76 | 0.05 | 0.05 | 0.69 | 0.83 | 1.00 | 11961.39 | 6184.18 |
| psi[5] | 0.54 | 0.54 | 0.04 | 0.04 | 0.49 | 0.61 | 1.00 | 8399.03 | 5926.44 |
| psi[6] | 0.51 | 0.50 | 0.03 | 0.03 | 0.45 | 0.56 | 1.00 | 8122.93 | 6418.99 |
| psi[7] | 0.69 | 0.68 | 0.04 | 0.04 | 0.62 | 0.75 | 1.00 | 12910.34 | 6676.54 |
| psi[8] | 0.48 | 0.48 | 0.03 | 0.03 | 0.43 | 0.53 | 1.00 | 9315.17 | 7043.50 |
| psi[9] | 0.78 | 0.78 | 0.05 | 0.04 | 0.71 | 0.86 | 1.00 | 11261.51 | 5939.97 |
| psi[10] | 0.84 | 0.84 | 0.05 | 0.05 | 0.77 | 0.92 | 1.00 | 13725.61 | 6430.39 |
| ObservedMean | variable | mean | median | sd | mad | q5 | q95 | rhat | ess_bulk | ess_tail | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| item1 | 2.367 | mu[1] | 2.363 | 2.363 | 0.086 | 0.086 | 2.222 | 2.504 | 1.003 | 1113.564 | 3020.860 |
| item2 | 1.955 | mu[2] | 1.951 | 1.953 | 0.084 | 0.083 | 1.810 | 2.088 | 1.005 | 687.119 | 2198.785 |
| item3 | 1.876 | mu[3] | 1.871 | 1.873 | 0.084 | 0.083 | 1.734 | 2.009 | 1.004 | 882.905 | 2490.044 |
| item4 | 2.011 | mu[4] | 2.008 | 2.007 | 0.084 | 0.084 | 1.870 | 2.145 | 1.005 | 782.884 | 2047.689 |
| item5 | 1.983 | mu[5] | 1.978 | 1.978 | 0.084 | 0.084 | 1.840 | 2.114 | 1.007 | 552.123 | 1622.828 |
| item6 | 1.893 | mu[6] | 1.888 | 1.888 | 0.076 | 0.077 | 1.763 | 2.014 | 1.007 | 599.939 | 1806.953 |
| item7 | 1.723 | mu[7] | 1.719 | 1.719 | 0.076 | 0.076 | 1.595 | 1.847 | 1.006 | 742.747 | 1914.120 |
| item8 | 1.842 | mu[8] | 1.838 | 1.838 | 0.072 | 0.072 | 1.718 | 1.956 | 1.007 | 585.199 | 1632.310 |
| item9 | 1.808 | mu[9] | 1.804 | 1.804 | 0.086 | 0.086 | 1.661 | 1.947 | 1.006 | 760.884 | 2237.888 |
| item10 | 1.520 | mu[10] | 1.517 | 1.517 | 0.081 | 0.080 | 1.386 | 1.651 | 1.003 | 1040.830 | 3151.001 |
Q1: The U.S. invasion of Iraq was not part of a campaign to fight terrorism, but was driven by oil companies and Jews in the U.S. and Israel.
Q1 has the highest agreement level for those with average conspiracy belief
At this point, one should investigate model fit of the model
If the model does not fit, then all model parameters could be biased
Moreover, the uncertainty accompanying each parameter (the posterior standard deviation) also be biased
But, to teach generalized measurement models, we will talk about differing models for observed data
Different distributions
Different parametrizations across different distributions
One plot that can help provide information about the item parameters is the item characteristic curve (ICC)
Not called this in Confirmatory Factor Analysis (but equivalent)
The ICC is the plot of the expected value of the response conditional on the value of the latent traits, for a range of latent trait values
E(Ypi∣θp)=μi+λiθp
ICC=f(E(Ypi∣θp),θp)
Because we have sampled values for each parameter, we can plot one ICC for each posterior draw
More results from conspiracy data
Compare results to blavaan package