Lecture 11: R Package
devtools package
Educational Statistics and Research Methods (ESRM) Program*
University of Arkansas
2025-03-19
Learning objectives
- Recognize the basic structure and purpose of an R package
- Create a simple R package skeleton using the devtools package
- Recognize the key directives in a NAMESPACE file
- Create R function documentation using roxygen2
- Create an R package that contains data (and associated documentation)
Prerequisite
- Before you start…
- If you are developing packages that contain only R code, then the tools you need come with R and RStudio.
- If you want to build packages with compiled C, C++, or Fortran code (or which to build other people’s packages with such code), then you will need to install additional tools.
- the Xcode development environment, which comes with the C compiler (
clang) - you need a Fortran compiler for older packages containing Fortran code. You can download the GNU Fortran Compiler from the R for Mac tools page.
- Rtools is a package to build R packages. The Rtools package comes in different versions, depending on the version of R that you are using.
- No other tools needed for developing R package on Linux/Unix.
R Package
The objectives of this section
- Recognize the basic structure and purpose of an R package
- Recognize the key directives in a NAMESPACE file
Basic Structure of R Package
- The two required sub-directories are:
R, which contains all of your R code filesman, which contains your documentation files.
- At the top level of your package directory, you will have
DESCRIPTIONfileNAMESPACEfile
- As an example, this is the file structure of the package
ESRM6990V:
DESCRIPTION file
The DESCRIPTION file contains key metadata for the package that is used by repositories like CRAN and by R itself.
In particular, this file contains the package name, the version number, the author and maintainer contact information, the license information, as well as any dependencies on other packages.
As an example, you can check ggplot2’s DESCRIPTION file on their github page.
Package: ggplot2
Title: Create Elegant Data Visualisations Using the Grammar of Graphics
Version: 3.5.1.9000
Authors@R: c(
person("Hadley", "Wickham", , "hadley@posit.co", role = "aut",
comment = c(ORCID = "0000-0003-4757-117X")),
person("Winston", "Chang", role = "aut",
comment = c(ORCID = "0000-0002-1576-2126")),
person("Lionel", "Henry", role = "aut"),
person("Thomas Lin", "Pedersen", , "thomas.pedersen@posit.co", role = c("aut", "cre"),
comment = c(ORCID = "0000-0002-5147-4711")),
person("Kohske", "Takahashi", role = "aut"),
person("Claus", "Wilke", role = "aut",
comment = c(ORCID = "0000-0002-7470-9261")),
person("Kara", "Woo", role = "aut",
comment = c(ORCID = "0000-0002-5125-4188")),
person("Hiroaki", "Yutani", role = "aut",
comment = c(ORCID = "0000-0002-3385-7233")),
person("Dewey", "Dunnington", role = "aut",
comment = c(ORCID = "0000-0002-9415-4582")),
person("Teun", "van den Brand", role = "aut",
comment = c(ORCID = "0000-0002-9335-7468")),
person("Posit, PBC", role = c("cph", "fnd"))
)
Description: A system for 'declaratively' creating graphics, based on "The
Grammar of Graphics". You provide the data, tell 'ggplot2' how to map
variables to aesthetics, what graphical primitives to use, and it
takes care of the details.
License: MIT + file LICENSE
URL: https://ggplot2.tidyverse.org, https://github.com/tidyverse/ggplot2
BugReports: https://github.com/tidyverse/ggplot2/issues
Depends:
R (>= 4.0)
Imports:
cli,NAMESPACE file
- The NAMESPACE file specifies:
- Exported function that is presented to the user. Functions that are not exported cannot be called directly by the user (although see below).
- What functions or packages are imported by the package.
In building R package, you don’t need to edit NAMESPACE file manually. You just write up your function by specifying which external functions you imported. document() function will automatically create/update NAMESPACE for you.
As an example, this is the NAMESPACE file for ESRM6990V (see GitHub). There is only one function jihong() existing that users can call in this package.
An example of mvtsplot package
export("mvtsplot")
import(splines)
import(RColorBrewer)
importFrom("grDevices", "colorRampPalette", "gray")
importFrom("graphics", "abline", "axis", "box", "image", "layout",
"lines", "par", "plot", "points", "segments", "strwidth",
"text", "Axis")
importFrom("stats", "complete.cases", "lm", "na.exclude", "predict",
"quantile")In this NAMESPACE file:
import(), simply takes a package name as an argument, and the interpretation is that all exported functions from that external package will be accessible to your packageimportFrom(), takes a package and a series of function names as arguments. This directive allows you to specify exactly which function you need from an external package. For example, this package imports thecolorRampPalette()andgray()functions from thegrDevicespackage.
Example of same function name from different packages
- You may find two R functions from different packages have same names.
For example, the commonly used dplyr package has a function named filter(), which is also the name of a function in the stats package.
- In R, every function has a full name, which includes the package namespace as part of the name. This format is along the lines of
We can use the following format to call these two functions to avoid confusion
R folder
The R sub-directory contains all of your R code, either in a single file, or in multiple files.
For larger packages it’s usually best to split code up into multiple files that logically group functions together.
The names of the R code files do not matter, but generally it’s not a good idea to have spaces in the file names.
As an example, I put the function jihong() inside of R/jihong.R code file.
jihong.R
#' This is the function for Jihong Zhang
#'
#' @param details Want to know more
#' @returns describe some basic information about Jihong Zhang
#' @examples
#' jihong(details = TRUE)
#' @export
jihong <- function(details = FALSE){
TEXT <- "Jihong Zhang is an Assistant Professor at University of Arkansas"
if (details == TRUE) {
TEXT <- "Jihong Zhang currently hold the position of Assistant Professor of Educational Statistics and Research Methods (ESRM) at the department of Counseling, Leadership, and Research Methods (CLRM), University of Arkansas. Previously, He served as a postdoctoral fellow at the Chinese University of Hong Kong in the department of Social Work. His academic journey in psychometrics starts with a doctoral training with Dr. Jonathan Templin in the Educational Measurement and Statistics (EMS) program at the University of Iowa. His primary research recently focuses on reliability and validation of psychological/psychometric network, Bayesian latent variable modeling, Item Response Theory modeling, and other advanced psychometric modeling. His expertise lies in the application of advanced statistical modeling in the fields of psychology and education, including multilevel modeling and structural equation modeling. His work is characterized by a commitment to enhancing the methodological understanding and application of statistics in educational research and beyond."
}
message(TEXT)
}man folder
The man sub-directory contains the documentation files for all of the exported objects of a package (e.g., help package).
With the development of the
roxygen2package, we no longer need to do that and can write the documentation directly into the R code files.
Left is starting with #' will be used to generate .Rd files that are part of help pages of function. Right is the help page of the function.

devtools package
There are two ways of creating a new package
You can also initialize an R package in RStudio by selecting “File” -> “New Project” -> “New Directory” -> “R Package”.
Or you can use
create()function indevtoolspackage
> devtools::create("~/Downloads/jeremy")
✔ Creating /Users/jihong/Downloads/jeremy/.
✔ Setting active project to "/Users/jihong/Downloads/jeremy".
✔ Creating R/.
✔ Writing DESCRIPTION.
Package: jeremy
Title: What the Package Does (One Line, Title Case)
Version: 0.0.0.9000
Authors@R (parsed):
* First Last <first.last@example.com> [aut, cre] (YOUR-ORCID-ID)
Description: What the package does (one paragraph).
License: `use_mit_license()`, `use_gpl3_license()` or friends to
pick a license
Encoding: UTF-8
Roxygen: list(markdown = TRUE)
RoxygenNote: 7.3.2.9000
✔ Writing NAMESPACE.
✔ Writing jeremy.Rproj.
✔ Adding "^jeremy\\.Rproj$" to .Rbuildignore.
✔ Adding ".Rproj.user" to .gitignore.
✔ Adding "^\\.Rproj\\.user$" to .Rbuildignore.
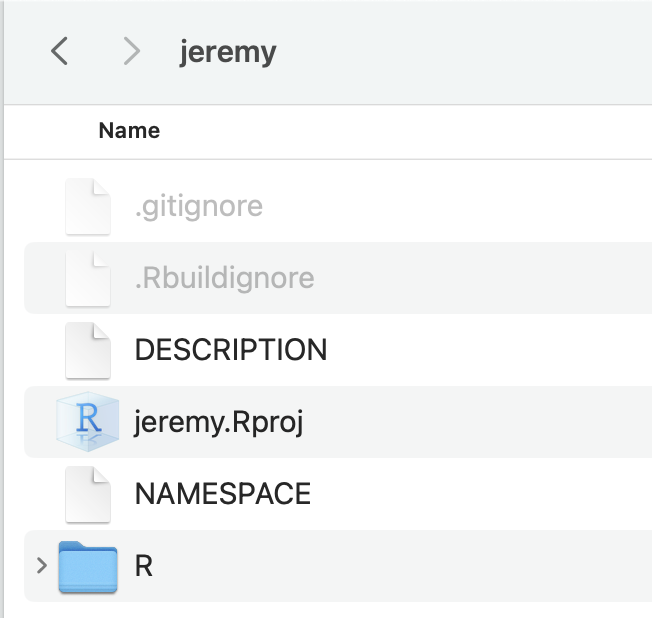
✔ Setting active project to "<no active project>".Below figure gives an example of what the new package directory will look like after you create an initial package structure with create or via the RStudio “New Project” interface.
- an R project file (
.Rprojextension) that saves some project options for the directory DESCRIPTIONandNAMESPACEfiles are automatically generated.gitignoreused to exclude some files in the directory by git.Rbuildignoreused to exclude some files when the package is built
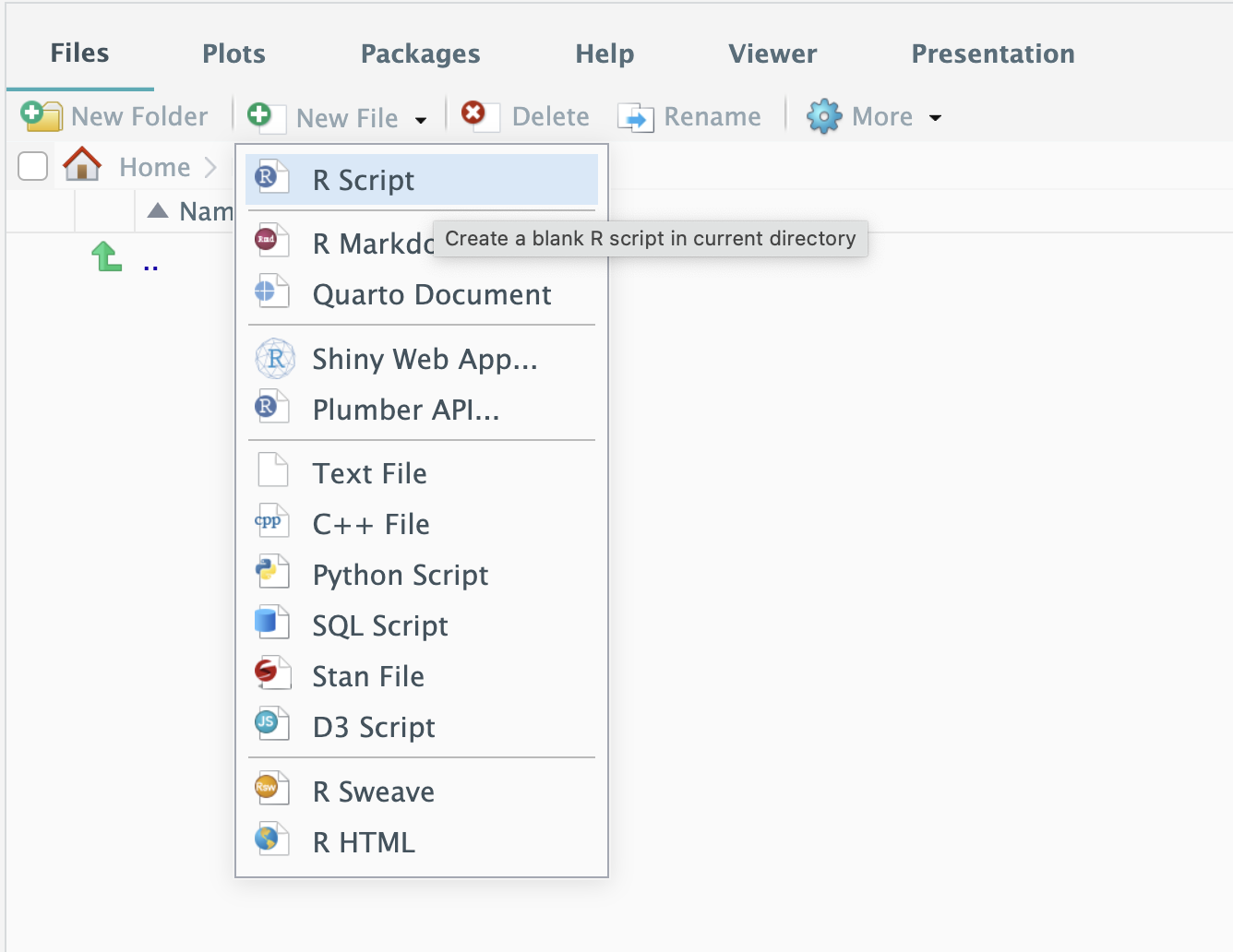
Example 1: Build your first R file in your first package
Open the .Rproj file will open Rstudio with your package directory as root path.
In your R folder, create a new R file named hello.R:
Exp1: Your first function — hello.R
Step 1: create the R file
Copy-and-paste the following starting template code into your hello.R file and save.
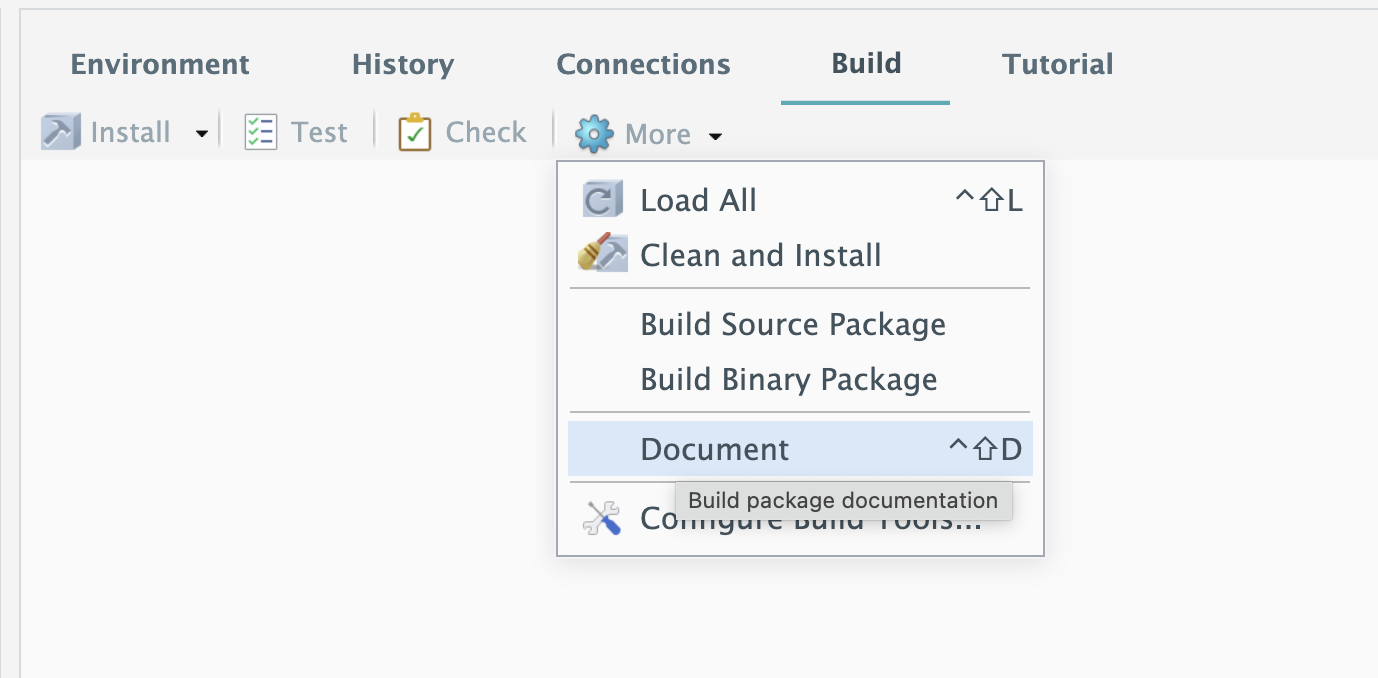
Step 2: document and load
Then, in Build panel, click More -> Document -> Load All.
These two buttons correspond to document() and load_all in devtools package, which are most frequently used.
Step 3: Test the function
Finally, in R console, type in hello(). Do you see the results?
Also, try adding question mark ? before the function to see the help page. Do you see your roxygen information works?
Finally, try to load your package. Did you see your package can be successfully loaded?
Help files: Procedure
To build help page of one function, we need to understand the procedure of R document building.
You put all of the help information (Title, description, Usage, Arguments, Value, Example) directly in the code where you define each function, with the format
#'.You call
document()or clickBuild>document, the roxygen2 package will convert the help information into.Rdfiles.These help files will ultimately be moved to a folder called
/manof your package, in an R documentation format (.Rd file extensions) that is fairly similar to LaTeX.
Help files: Tags
the
roxygen2package lets you put all of the help information directly in the code where you define each function. Further,roxygen2documentation allows you to include tags (@export,@importFrom) that will automate writing the package NAMESPACE file, so you don’t need to edit that file by hand.In our example, the following code starting with
@is called tags:
Some frequently used key tags:
@export: Export the function, so users will have direct access to it when they load the package@param: Explanation of a function parameter.@param [param_name] [explanation]@examples: Example code showing how to use the function@return: A description of the object returned by the function
You can find the full list of tags in roxygen2’s documentation (here).
Store Data in Package
If you want to store R objects and make them available to the user, put them in
data/. This is the best place to put example datasets. All the concrete examples above for data in a package and data as a package use this mechanism.If you want to store R objects for your own use as a developer, put them in
R/sysdata.rda. This is the best place to put internal data that your functions need.If you want to store data in some raw, non-R-specific form and make it available to the user, put it in
inst/extdata/.If you want to store dynamic data that reflects the internal state of your package within a single R session, use an environment. This technique is not as common or well-known as those above, but can be very useful in specific situations.
If you want to store data persistently across R sessions, such as configuration or user-specific data, use one of the officially sanctioned locations.
Example 1: Store your data into your package
- Step 4: Create Dataset.R
- Step 5: Create R script for data generation
- Step 5: run Dataset.R
- Step 6: Test the data
- Step 7: Share your package
In Dataset.R file, put the following R code into R file.
dat <- data.frame(
name = "jihong",
bio = "
I am a tenure-track Assistant Professor of Educational Statistics and Research Methods (ESRM) in the Department of Counseling, Leadership, and Research Methods (CLRM) at the University of Arkansas, Fayetteville, U.S.
Previously, I was a postdoctoral fellow in the Department of Social Work at the Chinese University of Hong Kong (CUHK). My academic journey in psychometrics began with doctoral training under Dr. Jonathan Templin in the Educational Measurement and Statistics (EMS) program at the University of Iowa. I have extensive experience in educational assessment. During my master’s program at the University of Kansas, I worked as a research assistant for three years in the Kansas Assessment Program (KAP), contributing to various assessment initiatives, including Dynamic Learning Maps (DLM). Later, during my Ph.D. program, I interned at the Stanford Research Institute (SRI), where I focused on digital learning in computerized testing.
My primary research interests include empirical and methodological studies in psychological network modeling, AI in Education, Bayesian latent variable modeling, Item Response Theory modeling, and other advanced psychometric methods. My expertise lies in applying advanced statistical modeling techniques in psychology and education.
"
)
usethis::use_data(dat, overwrite = TRUE)Open Dataset.R and Click the Source button to run the R file. You shall see the following output in R console.
>>> usethis::use_data(dat, overwrite = TRUE)
✔ Setting active project to "/Users/jihong/Downloads/jeremy".
✔ Adding R to Depends field in DESCRIPTION.
✔ Creating data/.
✔ Setting LazyData to "true" in DESCRIPTION.
✔ Saving "dat" to "data/dat.rda".
☐ Document your data (see <https://r-pkgs.org/data.html>).
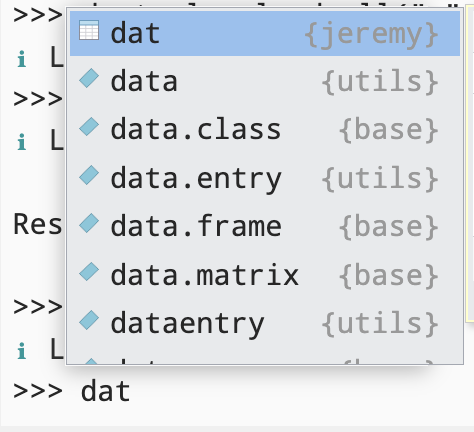
Type in the data name, such as dat in the R console. Do you see the data in your menu?
Currently, you can compress your package and share the zip file to anyone you want to share with.
As an example, download jeremy.zip on the webpage of Syllabus. Unzip it to Downloads folder and run the following R script.
install.packages("~/Downloads/jeremy", repos = NULL, type="source")
>> Installing package into ‘/Users/jihong/Rlibs’
>> (as ‘lib’ is unspecified)
>> * installing *source* package ‘jeremy’ ...
>> ** using staged installation
>> ** R
>> ** data
>> *** moving datasets to lazyload DB
>> ** byte-compile and prepare package for lazy loading
>> ** help
>> *** installing help indices
>> ** building package indices
>> ** testing if installed package can be loaded from temporary location
>> ** testing if installed package can be loaded from final location
>> ** testing if installed package keeps a record of temporary installation path
>> * DONE (jeremy)More about R Package
We’ve learnt how to build a simplest R package. However, today class is scratching the surface of R package. There are more things when you build your more complicated package. Some of which are:
- Create unit tests for an R package using the testthat package
- Categorize errors in the R CMD check process
- Recall the principles of open source software
- Recall two open source licenses
- Create create a GitHub repository for an R package
- Create an R package that is tested and deployed on Travis
- Create an R package that is tested and deployed on Appveyor
- Recognize characteristics of R packages that are not cross-platform
You can take a look at the Reference slide if you want to know more.
Reference
- Mastering Software Development in R — Chapter 3 Building R Packages
roxygen2documentation- R Packages (2e) by Hadley Wickham and Jennifer Bryan

ESRM 64503