Lecture 04: ANOVA Assumptions Checking
Experimental Design in Education
Educational Statistics and Research Methods (ESRM) Program*
University of Arkansas
2025-02-10
Class Outline
- Go through three assumptions of ANOVA and their checking statistics
- Post-hoc tests for more group comparisons
- Example: Intervention and Verbal Acquisition
- After-class Exercise: Effect of Sleep Duration on Cognitive Performance
Review ANOVA Procedure
- Set hypotheses:
- Null hypothesis (\(H_0\)): All group means are equal.
- Alternative hypothesis (\(H_A\)): At least one group mean differs.
- Determine statistical parameters:
- Significance level \(\alpha\)
- Degrees of freedom for between-group (\(df_b\)) and within-group (\(df_w\))
- Find the critical F-value.
- Compute test statistic:
- Calculate F-ratio based on between-group and within-group sum of squares.
- Compare results:
- Either compare \(F_{\text{obs}}\) with \(F_{\text{crit}}\) or \(p\)-value with \(\alpha\).
- If \(p < \alpha\), reject \(H_0\).
ANOVA Type I Error
The way to compare the means of multiple groups (i.e., more than two groups)
- Compared to multiple t-tests, the ANOVA does not inflate Type I error rates.
- For F-test in ANOVA, Type I error rate is not inflated (testing the \(H_0\) once a time)
- Type I error rate is inflated when we do multiple comparison (testing multiple \(H_0\)s given the same data)
- Family-wise error rate (FWER): probability of at least one Type I error occur = \(1 − (1 − \alpha)^c\) where c =number of tests. Frequently used FWER methods include Bonferroni or FDR.
- Compared to multiple t-tests, the ANOVA does not inflate Type I error rates.
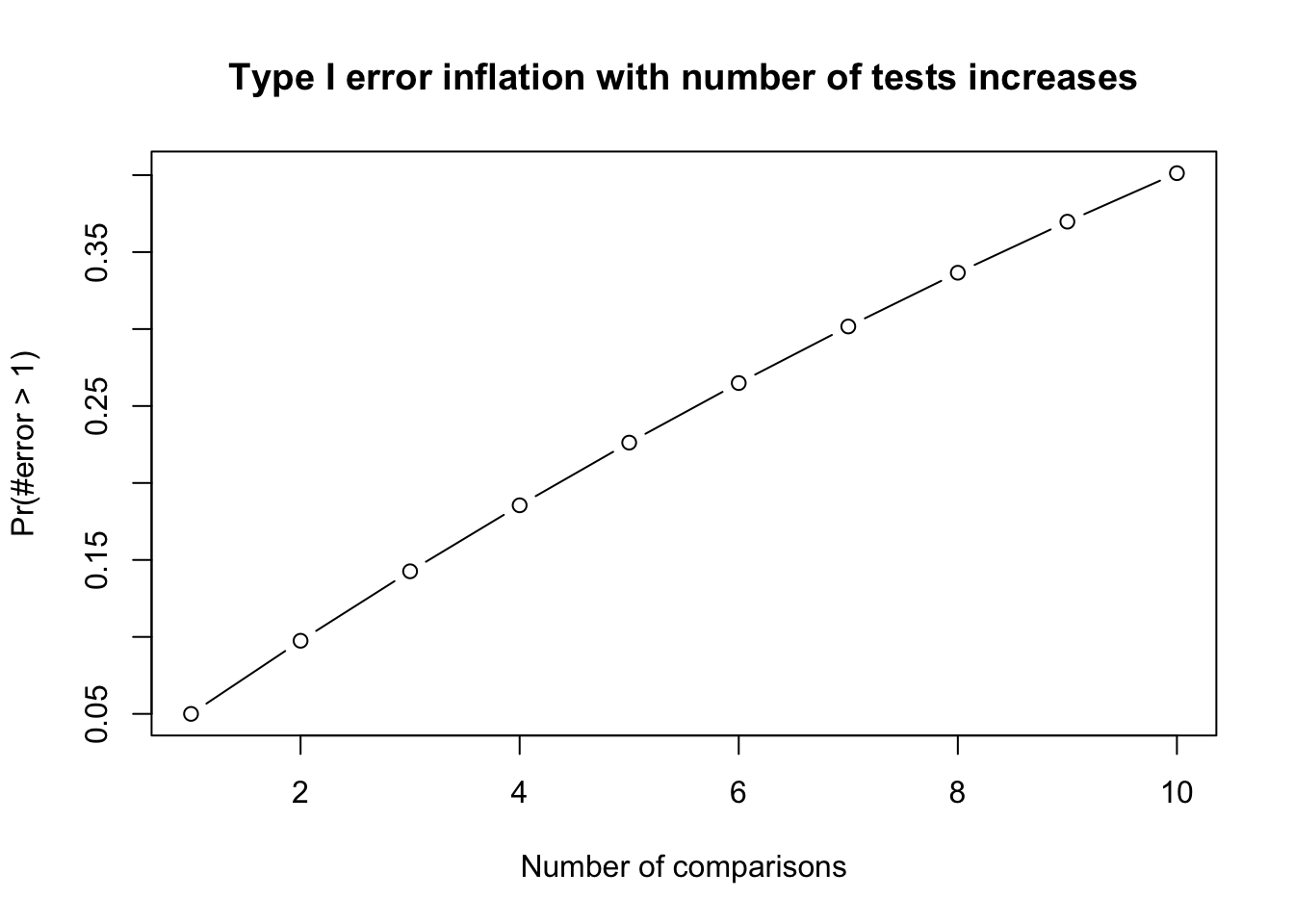
Family-Wise Error Rate
- The Family-Wise Error Rate (FWER) is the probability of making at least one Type I error (false positive) across a set of multiple comparisons.
- In Analysis of Variance (ANOVA), multiple comparisons are often necessary when analyzing the differences between group means. FWER control is crucial to ensure the validity of statistical conclusions.
1 ANOVA Assumptions
Overview
- Like all statistical tests, ANOVA requires certain assumptions to be met for valid conclusions:
- Independence: Observations are independent of each other.
- Normality: The residuals (errors) follow a normal distribution.
- Homogeneity of variance (HOV): The variance within each group is approximately equal.
Importance of Assumptions
- If assumptions are violated, the results of ANOVA may not be reliable.
- We call models robust when ANOVAs are not easily influenced by the violation of assumptions.
- Typically:
- ANOVA is robust to minor violations of normality, especially for large sample sizes (Central Limit Theorem).
- Not robust to violations of independence—if independence is violated, ANOVA is inappropriate.
- Moderately robust to HOV violations if sample sizes are equal.
Assumption 1: Independence among samples
- Definition: Each observation should be independent of others.
- Violations:
- Clustering of data (e.g., repeated measures).
- Participants influencing each other (e.g., classroom discussions).
- Check (Optional): Use the Durbin-Watson test to check Serial Correlation.
- Solution: Repeated Measures ANOVA, Mixed ANOVA, Multilevel Model
- Consequences: If independence is violated, ANOVA results are not valid.
Example: Demonstrating Independence Violation
Code
# Generate data with dependency (autocorrelation)
set.seed(1234)
n_per_group <- 20
groups <- 3
# Create dependent data using autoregressive structure
generate_dependent_data <- function(n, mean_val, sd_val, rho = 0.7) {
y <- numeric(n)
y[1] <- rnorm(1, mean_val, sd_val)
for(i in 2:n) {
y[i] <- rho * y[i-1] + rnorm(1, mean_val * (1-rho), sd_val * sqrt(1-rho^2))
}
return(y)
}
# Generate data for 3 groups with different means but with dependency
dependent_data <- data.frame(
group = factor(rep(c("A", "B", "C"), each = n_per_group)),
score = c(
generate_dependent_data(n_per_group, 50, 10), # Group A
generate_dependent_data(n_per_group, 55, 10), # Group B
generate_dependent_data(n_per_group, 60, 10) # Group C
)
)
# Compare with independent data
independent_data <- data.frame(
group = factor(rep(c("A", "B", "C"), each = n_per_group)),
score = c(
rnorm(n_per_group, 50, 10), # Group A
rnorm(n_per_group, 55, 10), # Group B
rnorm(n_per_group, 60, 10) # Group C
)
)
# Run ANOVA on both datasets
anova_dependent <- aov(score ~ group, data = dependent_data)
anova_independent <- aov(score ~ group, data = independent_data)
# Compare results
cat("ANOVA with DEPENDENT data:\n")
print(summary(anova_dependent))
cat("--------------------------\n")
cat("\nANOVA with INDEPENDENT data:\n")
print(summary(anova_independent))
# Check Durbin-Watson test
cat("--------------------------\n")
cat("\nDurbin-Watson test for dependent data:")
print(lmtest::dwtest(anova_dependent))
cat("--------------------------\n")
cat("\nDurbin-Watson test for independent data:")
print(lmtest::dwtest(anova_independent))ANOVA with DEPENDENT data:
Df Sum Sq Mean Sq F value Pr(>F)
group 2 695 347.3 3.72 0.0303 *
Residuals 57 5321 93.4
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
--------------------------
ANOVA with INDEPENDENT data:
Df Sum Sq Mean Sq F value Pr(>F)
group 2 377 188.55 2.389 0.101
Residuals 57 4500 78.94
--------------------------
Durbin-Watson test for dependent data:
Durbin-Watson test
data: anova_dependent
DW = 0.74297, p-value = 3.078e-09
alternative hypothesis: true autocorrelation is greater than 0
--------------------------
Durbin-Watson test for independent data:
Durbin-Watson test
data: anova_independent
DW = 1.8171, p-value = 0.1641
alternative hypothesis: true autocorrelation is greater than 0Key observations:
- Dependent data often shows artificially smaller p-values (inflated significance)
- Standard errors are underestimated when independence is violated
- Durbin-Watson test helps detect this violation (DW ≠ 2, p < 0.05)
Background: Durbin-Watson (DW) Test
Note
The Durbin-Watson test is primarily used for detecting autocorrelation in time-series data.
In the context of ANOVA with independent groups, residuals are generally assumed to be independent.
- It’s good practice to check this assumption, especially if there’s a reason to suspect potential autocorrelation.
Important
- Properties of DW Statistic:
- \(H_0\): Independence of residual is satisfied.
- Ranges from 0 to 4.
- A value around 2 suggests no autocorrelation.
- Values approaching 0 indicate positive autocorrelation.
- Values toward 4 suggest negative autocorrelation.
- P-value
- P-Value: A small p-value (typically < 0.05) indicates violation of independence
Real-world examples of independence violations:
- Dependent data scenarios:
- Repeated measures: Testing the same students multiple times (pre-test, mid-test, post-test)
- Classroom clusters: Students in the same classroom influence each other’s performance
- Time series: Daily test scores where yesterday’s performance affects today’s
- Spatial correlation: Schools in the same district sharing similar resources and outcomes
- Independent data scenarios:
- Random sampling: Selecting students from different schools across different regions
- Single measurement: Each participant tested only once with no interaction
- Controlled assignment: Participants randomly assigned to groups with no cross-contamination
Why this matters: When observations are dependent, they don’t provide as much unique information as independent observations. This leads to overconfident results (smaller standard errors and p-values).
Visualization of indepdence test
Code
library(ggplot2)
library(gridExtra)
# Use the same data from previous example
n_per_group <- 20
# Generate dependent and independent data
dependent_data <- data.frame(
group = factor(rep(c("A", "B", "C"), each = n_per_group)),
score = c(
generate_dependent_data(n_per_group, 50, 10),
generate_dependent_data(n_per_group, 55, 10),
generate_dependent_data(n_per_group, 60, 10)
),
observation = 1:(3*n_per_group),
type = "Dependent (AR)"
)
independent_data <- data.frame(
group = factor(rep(c("A", "B", "C"), each = n_per_group)),
score = c(
rnorm(n_per_group, 50, 10),
rnorm(n_per_group, 55, 10),
rnorm(n_per_group, 60, 10)
),
observation = 1:(3*n_per_group),
type = "Independent"
)
# Combine data
combined_data <- rbind(dependent_data, independent_data)
# Plot 1: Time series plot showing autocorrelation
p1 <- ggplot(combined_data, aes(x = observation, y = score, color = group)) +
geom_line(size = 0.8) + geom_point(size = 1.5) +
facet_wrap(~type, scales = "free") +
labs(title = "Time Series: Independence vs Dependence",
x = "Observation Order", y = "Score") +
theme_minimal() + theme(legend.position = "bottom")
# Plot 2: Lag plot to show autocorrelation
create_lag_data <- function(data) {
data$score_lag1 <- c(NA, data$score[-length(data$score)])
return(data[!is.na(data$score_lag1), ])
}
lag_dependent <- create_lag_data(dependent_data)
lag_independent <- create_lag_data(independent_data)
lag_combined <- rbind(lag_dependent, lag_independent)
p2 <- ggplot(lag_combined, aes(x = score_lag1, y = score)) +
geom_point(aes(color = group), alpha = 0.7) +
geom_smooth(method = "lm", se = TRUE, color = "black", size = 0.5) +
facet_wrap(~type, scales = "free") +
labs(title = "Lag Plot: Current vs Previous Observation",
x = "Score (t-1)", y = "Score (t)") +
theme_minimal() + theme(legend.position = "bottom")
# Display plots
grid.arrange(p1, p2, ncol = 1)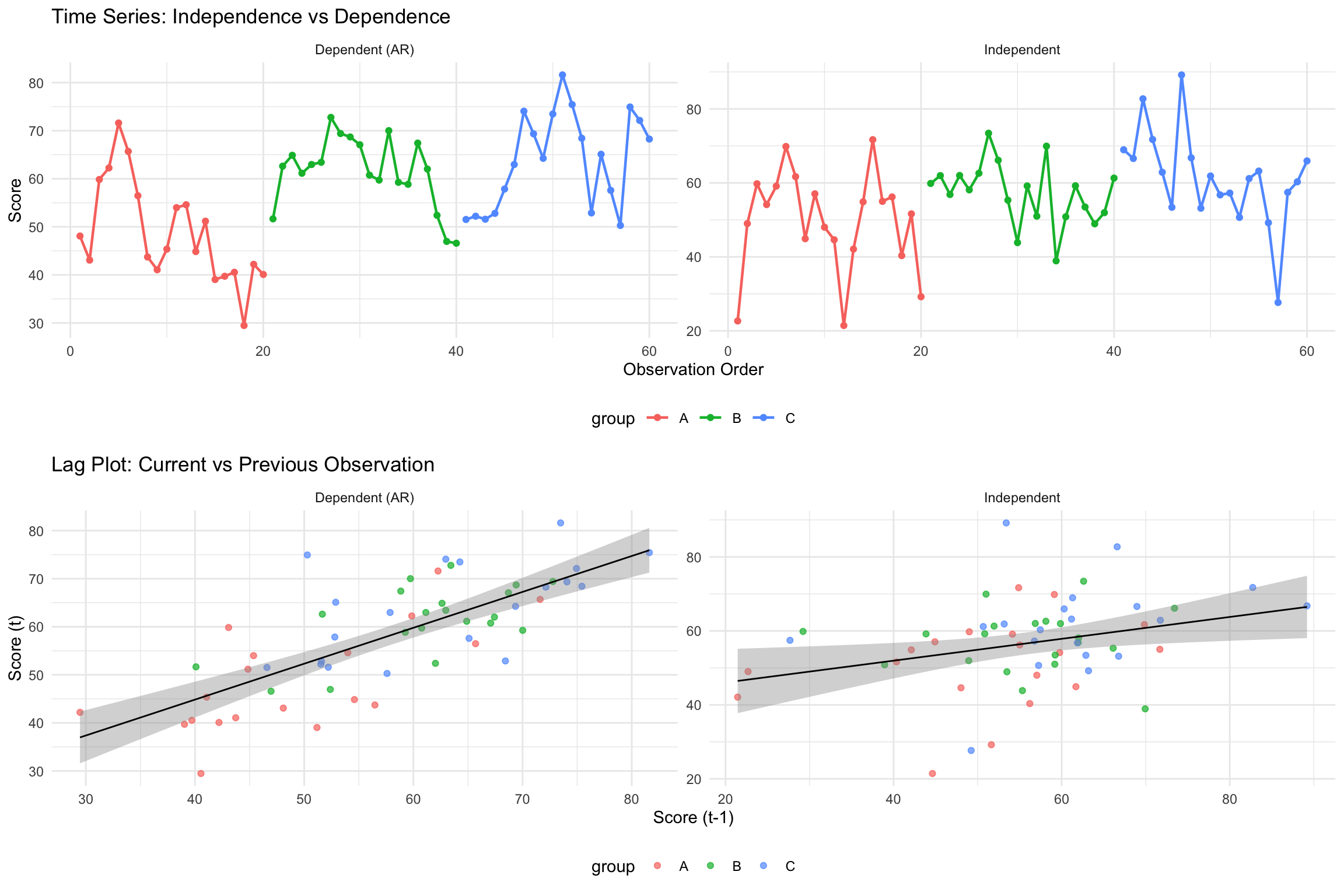
Key Visual Differences: - Dependent data: Shows smooth, connected patterns with visible trends - Independent data: Shows random scatter with no systematic patterns - Lag plot: Dependent data shows positive correlation between consecutive observations
lmtest::dwtest()
Performs the Durbin-Watson test for autocorrelation of disturbances.
The Durbin-Watson test has the null hypothesis that the autocorrelation of the disturbances is 0.
These results suggest there is no autocorrelation at the alpha level of .05.
Assumption 2: Normality
- The dependent variable (DV) should be normally distributed within each group.
- Assessments:
- Graphical methods: Histograms, Q-Q plots.
- Statistical tests:
- Shapiro-Wilk test (common)
- Kolmogorov-Smirnov (KS) test (for large samples)
- Anderson-Darling test (detects kurtosis issues).
- Robustness:
- ANOVA is robust to normality violations for large samples.
- If normality is violated, consider transformations or non-parametric tests.
Normality check for the whole samples
Assume that the DV (Y) is distributed normally within each group for ANOVA
ANOVA is robust to minor violations of normality
- So generally start with homogeneity, then assess normality
R function shapiro.test()
Code
set.seed(123) # For reproducibility
sample_normal_data <- rnorm(200, mean = 50, sd = 10) # Generate normal data
sample_nonnormal_data <- sample(1:10, 200, replace = TRUE) # Generate non-normal data
cat("Normality assumption holds:")
shapiro.test(sample_normal_data)
cat("Normality assumption does not hold:")
shapiro.test(sample_nonnormal_data)Normality assumption holds:
Shapiro-Wilk normality test
data: sample_normal_data
W = 0.99076, p-value = 0.2298
Normality assumption does not hold:
Shapiro-Wilk normality test
data: sample_nonnormal_data
W = 0.93581, p-value = 1.005e-07R function ks.test()
Warning in ks.test.default(scale(sample_nonnormal_data), "pnorm"): ties should
not be present for the one-sample Kolmogorov-Smirnov testNormality assumption holds:
Asymptotic one-sample Kolmogorov-Smirnov test
data: scale(sample_normal_data)
D = 0.054249, p-value = 0.5983
alternative hypothesis: two-sided
Normality assumption does not hold:
Asymptotic one-sample Kolmogorov-Smirnov test
data: scale(sample_nonnormal_data)
D = 0.12037, p-value = 0.006084
alternative hypothesis: two-sidedPerform Shapiro-Wilk and Kolmogorov-Smirnov for both datasets. Tell me which test works or does not work and why.
Normaliy check for each group
# Generate three-group data with normal distributions
library(dplyr) # we need this package to group the data
set.seed(123)
group_data <- data.frame(
group = rep(c("A", "B", "C"), each = 30),
score = c(rnorm(30, mean = 50, sd = 10), # Group A
rnorm(30, mean = 55, sd = 12), # Group B
rnorm(30, mean = 48, sd = 9)) # Group C
)
# Run Shapiro-Wilk test for each group
group_data |>
group_by(group) |>
summarize(
shapiro_p_value = shapiro.test(score)$p.value
)# A tibble: 3 × 2
group shapiro_p_value
<chr> <dbl>
1 A 0.797
2 B 0.961
3 C 0.848Interpretation of Normality Checking Results:
- If all p-values > 0.05: All groups satisfy normality assumption - proceed with ANOVA
- If one group has p-value < 0.05 but others are normal:
- ANOVA is generally robust to minor normality violations, especially with equal sample sizes
- Consider the sample size: larger samples (n > 30) make ANOVA more robust
- Options: (1) Continue with ANOVA if violation is mild, (2) Apply data transformation (log, square root), or (3) Use non-parametric alternative (Kruskal-Wallis test)
- If multiple groups violate normality: Strong evidence against using ANOVA - consider transformations or non-parametric tests
Perform Shapiro-Wilk for each group datasets. Tell me which group is not normal.
Assumption 3: Homogeneity of Variance (HOV)
- Variance across groups should be equal.
- Assessments:
- Levene’s test: Tests equality of variances.
- Brown-Forsythe test: More robust to non-normality.
- Bartlett’s test: For data with normality.
- Boxplots: Visual inspection.
- What if violated?
- Welch’s ANOVA (adjusted for variance differences).
- Transforming the dependent variable.
- Using non-parametric tests (e.g., Kruskal-Wallis).
Practical Considerations
## Computes Levene's test for homogeneity of variance across groups.
car::leveneTest(outcome ~ group, data = data)
## Boxplots to visualize the variance by groups
boxplot(outcome ~ group, data = data)
## Brown-Forsythe test
onewaytests::bf.test(outcome ~ group, data = data)
## Bartlett's Test
bartlett.test(outcome ~ group, data = data)- Levene’s test and Brown-Forsythe test are often preferred when data does not meet the assumption of normality, especially for small sample sizes.
- Bartlett’s test is most powerful when the data is normally distributed and when sample sizes are equal across groups. However, it can be less reliable if these assumptions are violated.
Decision Tree
ANOVA Robustness
Robust to:
- Minor normality violations (for large samples).
- Small HOV violations if group sizes are equal.
Not robust to:
- Independence violations—ANOVA is invalid if data points are dependent.
- Severe HOV violations—Type I error rates become unreliable.
The robustness of assumptions is something you should be careful about before/when you perform data collection. They are not something you can address after data collection has been finished.
Robustness to Violations of Normality Assumption
ANOVA assumes that the residuals (errors) are normally distributed within each group.
However, ANOVA is generally robust to violations of normality, particularly when the sample size is large.
Theoretical Justification: This robustness is primarily due to the Central Limit Theorem (CLT), which states that, for sufficiently large sample sizes (typically \(n≥30\) per group), the sampling distribution of the mean approaches normality, even if the underlying population distribution is non-normal.
This means that unless the data are heavily skewed or have extreme outliers, ANOVA results remain valid and Type I error rates are not severely inflated.
Robustness to Violations of Homogeneity of Variance
The homogeneity of variance (homoscedasticity) assumption states that all groups should have equal variances. ANOVA can tolerate moderate violations of this assumption, particularly when:
Sample sizes are equal (or nearly equal) across groups – When groups have equal sample sizes, the F-test remains robust to variance heterogeneity because the pooled variance estimate remains balanced.
The degree of variance heterogeneity is not extreme – If the largest group variance is no more than about four times the smallest variance, ANOVA results tend to remain accurate.
ANOVA: Lack of Robustness to Violations of Independence of Errors
The assumption of independence of errors means that observations within and between groups must be uncorrelated. Violations of this assumption severely compromise ANOVA’s validity because:
- Inflated Type I error rates – If errors are correlated (e.g., due to clustering or repeated measures), standard errors are underestimated, leading to an increased likelihood of falsely rejecting the null hypothesis.
- Biased parameter estimates – When observations are not independent, the variance estimates do not accurately reflect the true variability in the data, distorting F-statistics and p-values.
- Common sources of dependency – Examples include nested data (e.g., students within schools), repeated measurements on the same subjects, or time-series data. In such cases, alternatives like mixed-effects models or generalized estimating equations (GEE) should be considered.
2 Omnibus ANOVA Test
Overview
- What does it test?
- Whether there is at least one significant difference among means.
- Limitation:
- Does not tell which groups are different.
- Solution:
- Conduct post-hoc tests.
Individual Comparisons of Means
- If ANOVA is significant, follow-up tests identify where differences occur.
- Types:
- Planned comparisons: Defined before data collection.
- Unplanned (post-hoc) comparisons: Conducted after ANOVA.
Planned vs. Unplanned Comparisons
- Planned:
- Based on theory.
- Can be done even if ANOVA is not significant.
- Unplanned (post-hoc):
- Data-driven.
- Only performed if ANOVA is significant.
Types of Unplanned Comparisons
- Common post-hoc tests:
- Fisher’s LSD
- Bonferroni correction or Adjusted p-values
- Sidák correction
- Tukey’s HSD
Post-hoc test
Tukey’s HSD
- Controls for Type I error across multiple comparisons.
- Uses a q-statistic from a Tukey table.
- Preferred when all pairs need comparison.
Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = score ~ group, data = group_data)
$group
diff lwr upr p adj
B-A 7.611098 1.902128 13.320067 0.0057521
C-A -1.309179 -7.018149 4.399791 0.8483773
C-B -8.920277 -14.629246 -3.211307 0.0009985Family-wise Error Rate (adjusted p-values)
- Adjusts p-values to reduce Type I error
- Report adjusted p-values (typically larger that original p-values)
Tukey Honest Significant Differences (HSD) Test
Purpose: Conducts all possible pairwise comparisons between group means while controlling the family-wise error rate at the specified alpha level (typically 0.05).
Why it’s needed: Simple pairwise t-tests after a significant ANOVA inflate the Type I error rate. Tukey HSD adjusts for multiple comparisons to maintain the overall error rate.
How it works: Creates confidence intervals for all pairwise mean differences using the studentized range distribution, which accounts for the number of groups being compared.
Interpretation: If a confidence interval does not include zero, or if p adj < 0.05, the pair of groups differs significantly.
diff lwr upr p adj
B-A 7.611098 1.902128 13.320067 0.0057520929
C-A -1.309179 -7.018149 4.399791 0.8483772504
C-B -8.920277 -14.629246 -3.211307 0.0009984894Interpretation: The Tukey HSD test provides pairwise comparisons between all groups while controlling for family-wise error rate. The output shows:
- diff: The difference in means between each pair of groups
- lwr & upr: The 95% confidence interval for the difference
- p adj: The adjusted p-value for multiple comparisons
Pairs with p adj < 0.05 indicate statistically significant differences between those groups.
Interpreting Results
ANOVA Statistics
Df Sum Sq Mean Sq F value Pr(>F)
group 3 724.1 241.37 11.45 2.04e-05 ***
Residuals 36 759.2 21.09
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1- A one-way analysis of variance (ANOVA) was conducted to examine the effect of teaching method on students’ test scores. The results indicated a statistically significant difference in test scores across the four groups, (F(3, 36) = 11.45, p < .001). Post-hoc comparisons using the Tukey HSD test revealed that the Interactive method (G3) (M = 28.06, SD = 3.33) resulted in significantly higher scores than the Traditional method (M = 18.17, SD = 4.47) with p < .001. However, no significant difference was found between G1 and the Control group (p = .99). These results suggest that certain teaching methods can significantly improve student performance compared to traditional approaches.
ANOVA Example: Intervention and Verbal Acquisition
Background
- Research Question: Does an intensive intervention improve students’ verbal acquisition scores?
- Study Design:
- 4 groups: Control, G1, G2, G3 (treatment levels).
- Outcome variable: Verbal acquisition score (average of three assessments).
- Hypotheses:
- \(H_0\): No difference in verbal acquisition scores across groups.
- \(H_A\): At least one group has a significantly different mean.
Step 1: Generate Simulated Data in R
# Load necessary libraries
library(tidyverse)
# Set seed for reproducibility
set.seed(123)
# Generate synthetic data for 4 groups
data <- tibble(
group = rep(c("Control", "G1", "G2", "G3"), each = 30),
verbal_score = c(
rnorm(30, mean = 70, sd = 10), # Control group
rnorm(30, mean = 75, sd = 12), # G1
rnorm(30, mean = 80, sd = 10), # G2
rnorm(30, mean = 85, sd = 8) # G3
)
)
# View first few rows
head(data)# A tibble: 6 × 2
group verbal_score
<chr> <dbl>
1 Control 64.4
2 Control 67.7
3 Control 85.6
4 Control 70.7
5 Control 71.3
6 Control 87.2Function explanations:
library(tidyverse): Loads the tidyverse package collection, including dplyr, ggplot2, and tibbleset.seed(123): Sets the random number generator seed to ensure reproducible results across runstibble(): Creates a modern data frame (tibble) with improved printing and subsetting behaviorrep(c("Control", "G1", "G2", "G3"), each = 30): Repeats each group label 30 times, creating 120 total observations (30 per group)rnorm(30, mean = X, sd = Y): Generates 30 random numbers from a normal distribution with specified mean and standard deviationc(): Combines the four vectors of random numbers into a single vector of 120 valueshead(data): Displays the first 6 rows of the dataset for preview
Step 2: Summary Statistics
# A tibble: 4 × 4
group mean_score sd_score n
<chr> <dbl> <dbl> <int>
1 Control 69.5 9.81 30
2 G1 77.1 10.0 30
3 G2 80.2 8.70 30
4 G3 84.2 7.25 30Function explanations:
group_by(group): Groups the data by thegroupvariable, allowing subsequent operations to be performed separately for each groupsummarise(): Creates summary statistics for each group, collapsing the grouped data into summary valuesmean(verbal_score): Calculates the arithmetic mean of verbal scores within each groupsd(verbal_score): Computes the standard deviation of verbal scores within each groupn(): Counts the number of observations in each group
# Boxplot visualization
library(ggplot2) # Load ggplot2 package for creating visualizations
ggplot(data, aes(x = group, y = verbal_score, fill = group)) + # Create base plot with group on x-axis, scores on y-axis, fill by group
geom_boxplot() + # Add boxplot layer to show distribution of scores within each group
theme_minimal() + # Apply minimal theme for clean appearance
labs(title = "Verbal Acquisition Scores Across Groups", y = "Score", x = "Group") # Add descriptive labels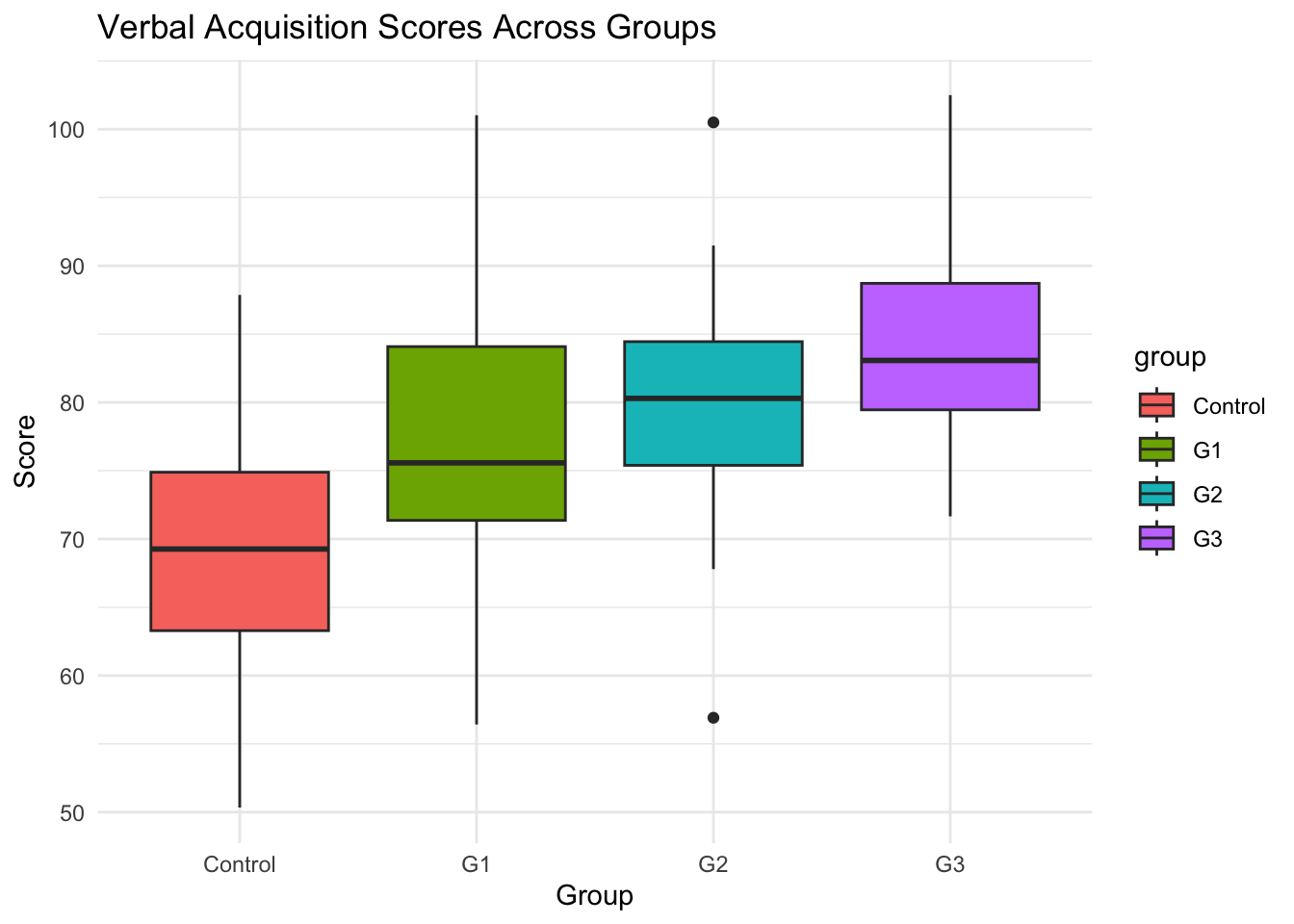
Step 3: Check ANOVA Assumptions
Assumption Check 1: Independence of residuals Check
Durbin-Watson test
data: anova_model
DW = 2.0519, p-value = 0.5042
alternative hypothesis: true autocorrelation is greater than 0- Interpretation:
- In this example, the
DWvalue is close to 2, and the p-value is greater than 0.05, indicating no significant autocorrelation in the residuals.
- In this example, the
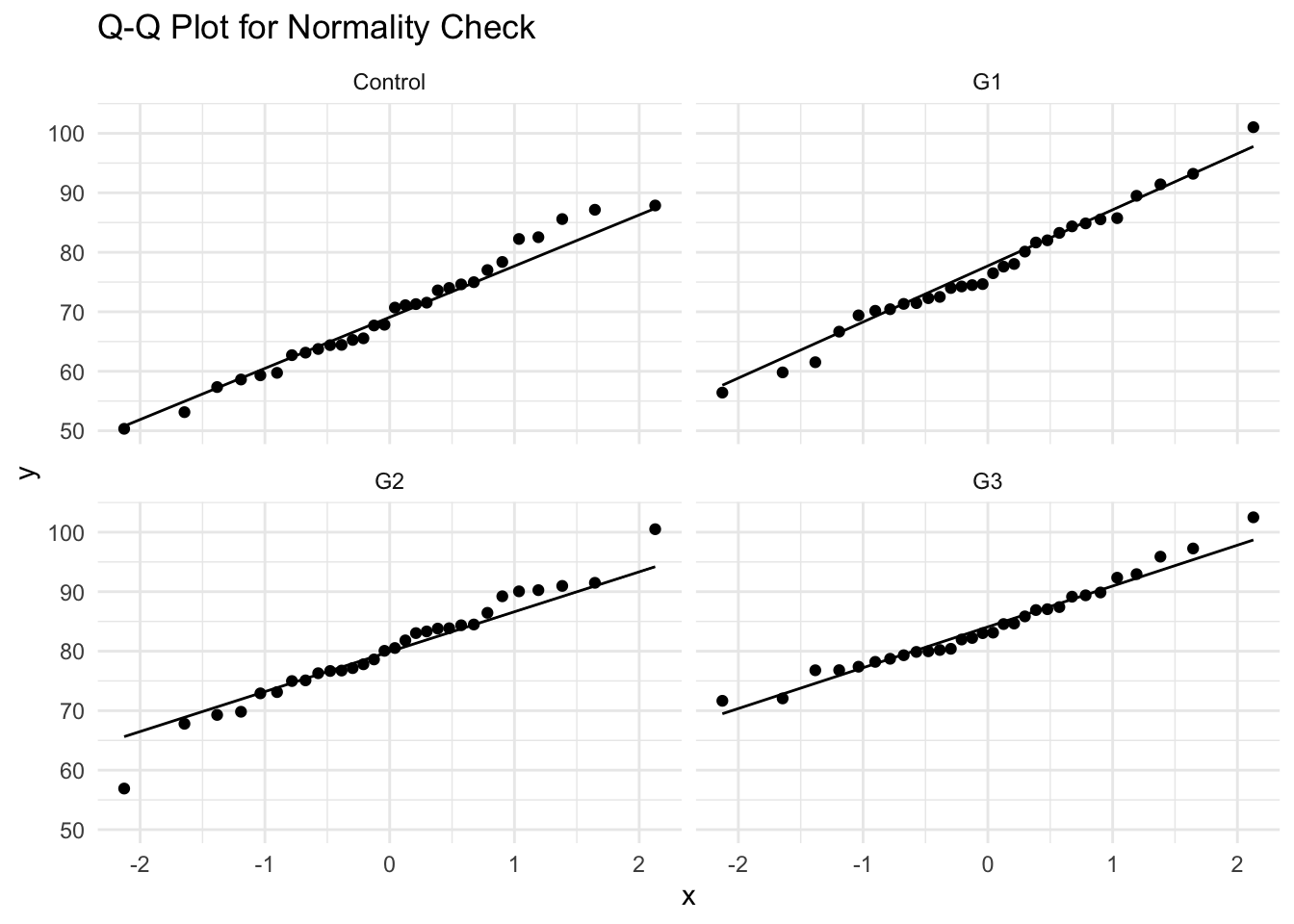
Assumption Check 2: Normality Check
# A tibble: 4 × 2
group shapiro_p
<chr> <dbl>
1 Control 0.797
2 G1 0.961
3 G2 0.848
4 G3 0.568- Interpretation:
- If \(p>0.05\), normality assumption is not violated.
- If \(p<0.05\), data deviates from normal distribution.
- Since the data meets the normality requirement and no outliers, we can use Bartlett’s or Levene’s tests for HOV checking.
- Alternative Check: Q-Q Plot
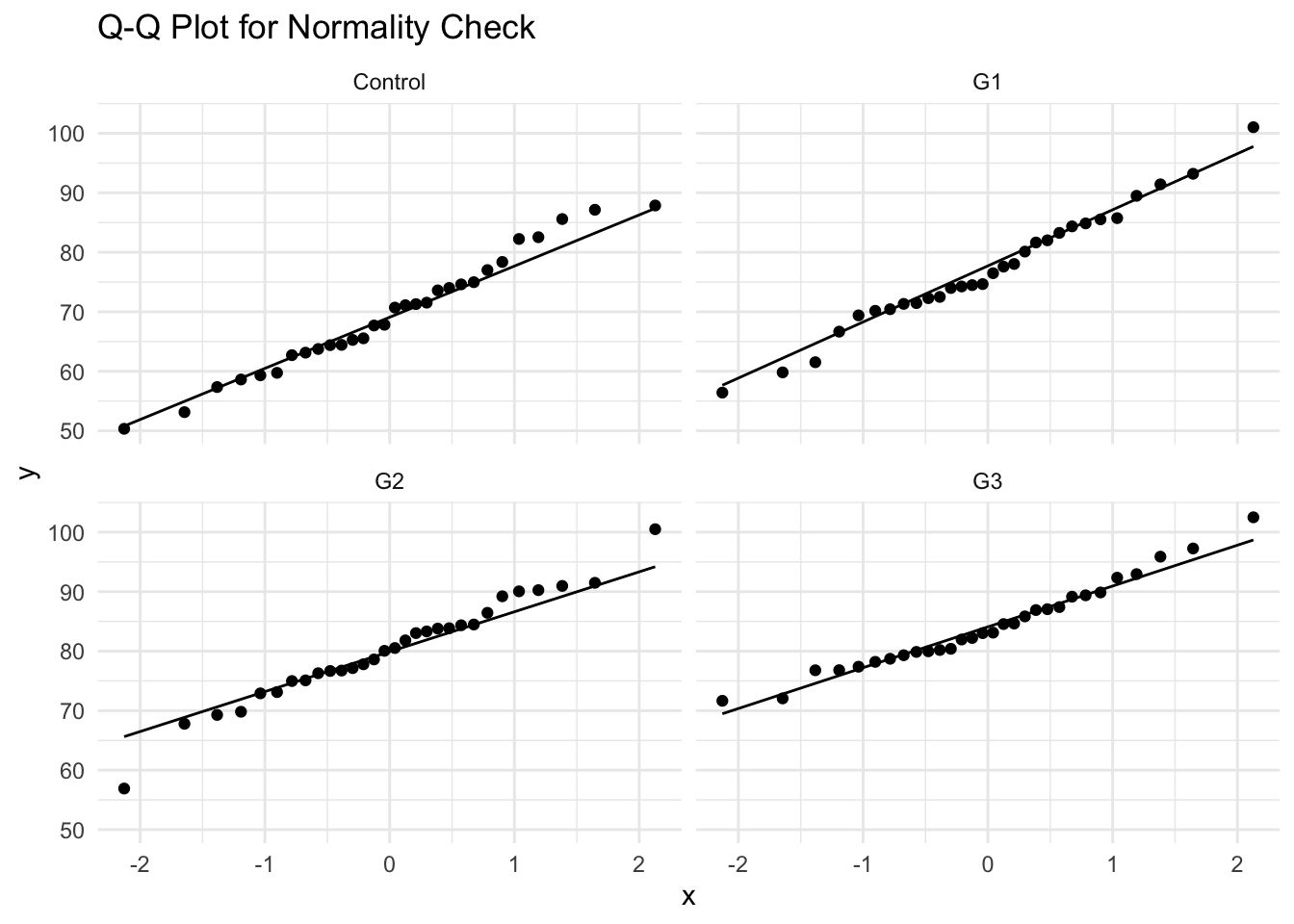
Assumption Check 3: Homogeneity of Variance (HOV) Check
library(car) # for leveneTest
library(onewaytests) # for bf.test
data$group <- as.factor(data$group)
car::leveneTest(verbal_score ~ group, data = data)
cat("-----------------------------\n")
bartlett.test(verbal_score ~ group, data = data)
cat("-----------------------------\n")
onewaytests::bf.test(verbal_score ~ group, data = data)Levene's Test for Homogeneity of Variance (center = median)
Df F value Pr(>F)
group 3 1.1718 0.3236
116
-----------------------------
Bartlett test of homogeneity of variances
data: verbal_score by group
Bartlett's K-squared = 3.5385, df = 3, p-value = 0.3158
-----------------------------
Brown-Forsythe Test (alpha = 0.05)
-------------------------------------------------------------
data : verbal_score and group
statistic : 14.32875
num df : 3
denom df : 109.9888
p.value : 6.030497e-08
Result : Difference is statistically significant.
------------------------------------------------------------- - Interpretation:
- If \(p>0.05\), variance is homogeneous (ANOVA assumption met).
- If \(p<0.05\), variance differs across groups (consider Welch’s ANOVA).
- It turns out our data does not violate the homogeneity of variance assumption.
Step 4: Perform One-Way ANOVA
Df Sum Sq Mean Sq F value Pr(>F)
group 3 3492 1164.1 14.33 5.28e-08 ***
Residuals 116 9424 81.2
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1- Interpretation:
- If \(p<0.05\), at least one group mean is significantly different.
- If \(p>0.05\), fail to reject \(H0\) (no significant differences).
Step 5: Post-Hoc Tests (Tukey’s HSD)
After finding a significant ANOVA result, we perform Tukey’s HSD test to identify which specific groups differ from each other.
For example, we can compare:
- G1 vs Control
- G2 vs Control
- G3 vs Control
- G2 vs G1
- G3 vs G1
- G3 vs G2
diff lwr upr p adj
G1-Control 7.611 1.545 13.677 0.008
G2-Control 10.715 4.649 16.782 0.000
G3-Control 14.720 8.654 20.786 0.000
G2-G1 3.104 -2.962 9.170 0.543
G3-G1 7.109 1.042 13.175 0.015
G3-G2 4.005 -2.062 10.071 0.318- Interpretation:
- Identifies which groups differ.
- If \(p<0.05\), the groups significantly differ.
- G1-Control
- G2-Control
- G3-Control
- G3-G1
Step 6: Family-wise Error Rate Control
General Linear Hypothesis Tests:
glhtfunction in multcomp package allow us to perform family-wise error rate control (usingadjusted p-values).Alternatively, we can use
TukeyHSDto adjust p values to control inflated Type I error.
\[ Y = \beta_0G_0+\beta_1G_1 +\beta_2G_2 +\beta_3G_3 \]
- When \(\{G_0, G_1, G_2, G_3\}\) = \(\{0, 1, 0, 0\}\), \(Y = \beta_1\), which is the group difference between G1 with the control group
- When \(\{G_0, G_1, G_2, G_3\}\) = \(\{0, 0, 1, 0\}\), \(Y = \beta_2\), which is the group difference between G2 with the control group
- When \(\{G_0, G_1, G_2, G_3\}\) = \(\{0, -1, 1, 0\}\), \(Y = \beta_2 - \beta_1\), which is the group difference between G2 with G1
library(multcomp)
# install.packages("multcomp")
### set up multiple comparisons object for all-pair comparisons
# head(model.matrix(anova_model))
comprs <- rbind(
"G1 - Ctrl" = c(0, 1, 0, 0),
"G2 - Ctrl" = c(0, 0, 1, 0),
"G3 - Ctrl" = c(0, 0, 0, 1),
"G2 - G1" = c(0, -1, 1, 0),
"G3 - G1" = c(0, -1, 0, 1),
"G3 - G2" = c(0, 0, -1, 1)
)
cht <- glht(anova_model, linfct = comprs)
summary(cht, test = adjusted("fdr"))
Simultaneous Tests for General Linear Hypotheses
Fit: aov(formula = verbal_score ~ group, data = data)
Linear Hypotheses:
Estimate Std. Error t value Pr(>|t|)
G1 - Ctrl == 0 7.611 2.327 3.270 0.00283 **
G2 - Ctrl == 0 10.715 2.327 4.604 3.20e-05 ***
G3 - Ctrl == 0 14.720 2.327 6.325 2.94e-08 ***
G2 - G1 == 0 3.104 2.327 1.334 0.18487
G3 - G1 == 0 7.109 2.327 3.055 0.00419 **
G3 - G2 == 0 4.005 2.327 1.721 0.10555
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Adjusted p values reported -- fdr method)Side-by-Side comparison of two methods
TukeyHSD method
FDR method
- The differences in p-value adjustment between the
TukeyHSDmethod and themultcomppackage stem from how each approach calculates and applies the multiple comparisons correction. Below is a detailed explanation of these differences.
Comparison of Differences
| Feature | TukeyHSD() (Base R) |
multcomp::glht() |
|---|---|---|
| Distribution Used | Studentized Range (q-distribution) | t-distribution |
| Error Rate Control | Strong FWER control | Flexible error control |
| Simultaneous Confidence Intervals | Yes | Typically not (depends on method used) |
| Adjustment Method | Tukey-Kramer adjustment | Single-step, Westfall, Holm, Bonferroni, etc. |
| P-value Differences | More conservative (larger p-values) | Slightly different due to t-distribution |
Step 6: Reporting ANOVA Results
Result
We first examined three assumptions of ANOVA for our data as the preliminary analysis. According to the Durbin-Watson test, the Shapiro-Wilk normality test, and the Bartletts’ test, the sample data meets all assumptions of the one-way ANOVA modeling.
A one-way ANOVA was then conducted to examine the effect of three intensive intervention methods (Control, G1, G2, G3) on verbal acquisition scores. There was a statistically significant difference between groups, \(F(3,116)=14.33\), \(p<.001\).
To further examine which intervention method is most effective, we performed Tukey’s post-hoc comparisons. The results revealed that all three intervention methods have significantly higher scores than the control group (G1-Ctrl: p = .003; G2-Ctrl: p < .001; G3-Ctrl: p < .001). Among the three intervention methods, G3 seems to be the most effective. Specifically, G3 showed significantly higher scores than G1 (p = .004). However, no significant difference was found between G2 and G3 (p = .105).
Discussion
These findings suggest that higher intervention intensity improves verbal acquisition performance, which is consistent with prior literature [xxxx/references].
3 After-Class Exercise: Effect of Sleep Duration on Cognitive Performance
Background
Research Question:
- Does the amount of sleep affect cognitive performance on a standardized test?
Study Design
- Independent variable: Sleep duration (3 groups: Short (≤5 hrs), Moderate (6-7 hrs), Long (≥8 hrs)).
- Dependent variable: Cognitive performance scores (measured as test scores out of 100).
A one-way ANOVA was conducted to examine the effect of sleep duration on cognitive performance.
There was a statistically significant difference in cognitive test scores across sleep groups, \(F(2,87)=15.88\),\(p<.001\).
Tukey’s post-hoc test revealed that participants in the Long sleep group (M=81.52, SD=6.27) performed significantly better than those in the Short sleep group (M=65.68, SD=12.55), p<.01.
These results suggest that inadequate sleep is associated with lower cognitive performance.

ESRM 64503