---
title: "Reliability and Replicability of the Psychological Network Analysis"
date: 'May 3 2024'
categories:
- R
- Bayesian
- Tutorial
- Network Analysis
execute:
eval: true
echo: true
warning: false
fig-align: center
format:
html:
code-fold: true
code-summary: 'Click to see the code'
toc-location: left
toc-depth: 2
toc-expand: true
bibliography: references.bib
---
# Empirical study 1: PTSD networks
This study examines the reliability and replicability of the network structure of PTSD symptoms in a large sample of trauma-exposed individuals.
Fried et al. (2018) shared their correlation matrices, model output, and code in the [Supplementary Materials](https://doi.org/10.1080/00273171.2019.1616526) and encouraged reanalysis of the data for further replicability research.
This data sets were reanalyzed by [@forbesQuantifyingReliabilityReplicability2021].
## Research Plan
First, the PTSD network structure will be estimated through three models:
- **Model 1**: Gaussian graphical models (GGMs) derived from polychoric correlations using fused graphical lasso selecting tuning parameters using k-fold cross-validation first.
- **Model 2**: GGM using FIML in *psychonetrics* (`FLML_prune`)
- **Model 3**: Bayesian GGM using `BGGM_estimation`, which have best performance in sensitivity and specificity.
Second, the parameter estimation and network characteristics will be carefully screened and reviewed.
- Network characteristics across waves
1. Node strength (NS_max, NS_min)
2. Number of total possible edges (Num_total_edges)
3. Number of estimated (non-zero) edges (Num_nonzero_edges)
4. Connectivity -- percentage of edges that are non-zero (Connectivity)
5. Number of zero edges (Num_zero_edges)
6. Number of positive edges (Num_positive_edges)
7. Number of negative edges (Num_negative_edges)
- Bootstrap and NCT results
1. Edges bootstrapped CIs
2. Strength centrality bootstrapped CIs
3. NCT omnibus test
```{r}
##Network characteristics
network_char <- function(network) {
graph <- network$graph
#edge lists
edges <- graph[upper.tri(graph)]
#node strength
strengthT1<-centrality(graph)$OutDegree
NS_sum <- sum(strengthT1)
NS_max <- max(strengthT1)
NS_min <- min(strengthT1)
# Number of total possible edges
totalT1 <- length(edges)
Num_total_edges <- totalT1
# Number of estimated edges:
nedges<- sum(edges!=0)
Num_nonzero_edges <- nedges
#Connectivity
connectT1<-round(nedges/totalT1 * 100, 1)
Connectivity <- connectT1
#Number of zero edges
n0edges<- sum(edges==0)
Num_zero_edges <- n0edges
#Number of positive edges
nposedges<- sum(edges>0)
Num_positive_edges <- nposedges
#Number of negative edges
nnegedges<- sum(edges<0)
Num_negative_edges <- nnegedges
#Bootstrap confidence intervals
boot1a <- bootnet(network, nCores = 6, nBoots = 1000)
boot1a_summary <- summary(boot1a, type = "CIs", nCores = 6)
Num_nonzero_edges_bootstrap <- sum(sign(boot1a_summary$CIlower) == sign(boot1a_summary$CIupper) & sign(boot1a_summary$CIlower) != 0)
Num_positive_edges_bootstrap <- sum(sign(boot1a_summary$CIlower) == sign(boot1a_summary$CIupper) & sign(boot1a_summary$CIlower) == 1)
c(NS_sum = round(NS_sum, 2),
NS_max = round(NS_max, 2),
NS_min = round(NS_min, 2),
Connectivity= Connectivity,
Num_total_edges= Num_total_edges,
Num_nonzero_edges= Num_nonzero_edges,
Num_nonzero_edges_bootstrap = Num_nonzero_edges_bootstrap,
Num_zero_edges= Num_zero_edges,
Num_positive_edges= Num_positive_edges,
Num_positive_edges_bootstrap= Num_positive_edges_bootstrap,
Num_negative_edges= Num_negative_edges)
}
```
## Data Information
"*The primary analyses were based on a subset of community participants from a larger longitudinal study. Analyses included 403 participants who completed questions online regarding depression and anxiety symptoms two times one week apart.* [@forbesQuantifyingReliabilityReplicability2021]"
| Node label | Symptom |
|------------|-------------------------------------------------------------------------|
| PHQ1 | Little interest or pleasure in doing things |
| PHQ2 | Feeling down, depressed, or hopeless |
| PHQ3 | Trouble falling or staying asleep, or sleeping too much |
| PHQ4 | Feeling tired or having little energy |
| PHQ5 | Poor appetite or overeating |
| PHQ6 | Feeling bad about yourself — or that you are a failure or have let yourself or your family down |
| PHQ7 | Trouble concentrating on things, such as reading the newspaper or watching television |
| PHQ8 | Moving or speaking so slowly that other people could have noticed? Or the opposite—being so fidgety or restless that you have been moving around a lot more than usual |
| PHQ9 | Thoughts that you would be better off dead or of hurting yourself in some way |
| GAD1 | Feeling nervous, anxious, or on edge |
| GAD2 | Not being able to stop or control worrying |
| GAD3 | Worrying too much about different things |
| GAD4 | Trouble relaxing |
| GAD5 | Being so restless that it’s hard to sit still |
| GAD6 | Becoming easily annoyed or irritable |
| GAD7 | Feeling afraid as if something awful might happen |
## Data Analysis
To replicate the same analysis as @forbesQuantifyingReliabilityReplicability2021, we used the raw data from their OSF project (<https://osf.io/6fk3v/>)
```{r}
#| code-summary: "List the files in the OSF project of (Forbes et al., 2021)"
library(osfr)
forbes2018 <- project <- osf_retrieve_node("https://osf.io/6fk3v/")
osf_ls_files(forbes2018)
```
Download the two-wave data set (`time1and2data_wide.csv`).
```{r}
#| eval: false
#| code-summary: "Download the time1and2data_wide.csv"
forbes2018_files <- osf_ls_files(forbes2018)
osf_download(forbes2018_files[7, ], path = "PTSD_network/")
osf_download(forbes2018_files[4, ], path = "PTSD_network/")
```
There are 403 participants with 40 variables and 1 ID.
```{r}
#| code-summary: "Read in the data set"
# Set the random seed:
set.seed(123)
# Load time1 dataset:
time1and2data <- read.csv("PTSD_network/time1and2data_wide.csv", header=FALSE)
time1and2data<-as.data.frame(time1and2data)
time1data<- time1and2data[c(2:17)]
time1data<-as.data.frame(time1data)
labels_time1and2data <- c("PHQ1","PHQ2","PHQ3","PHQ4","PHQ5","PHQ6","PHQ7",
"PHQ8","PHQ9","GAD1","GAD2","GAD3","GAD4","GAD5",
"GAD6","GAD7")
groups_time1and2data <- c(rep("Depression", 9), rep("Anxiety", 7))
time2data<- time1and2data[c(18:33)]
time2data<-as.data.frame(time2data)
colnames(time2data) <- colnames(time1data) <- labels_time1and2data
Labels<- c("interest", "feel down", "sleep", "tired", "appetite", "self-esteem",
"concentration", "psychomotor", "suicidality", "feel nervous",
"uncontrollable worry", "worry a lot", "trouble relax", "restless",
"irritable", "something awful")
time1dep<-time1data[c(1:9)]
time1anx<-time1data[c(10:16)]
time2dep<-time2data[c(1:9)]
time2anx<-time2data[c(10:16)]
```
The Patient Health Questionnaire (PHQ-9; Kroenke, Spitzer & Williams, 2001) is a 9-item measure of depression symptoms with cutoff scores that indicate clinically significant levels of major depression.
The Brief Measure for Assessing Generalized Anxiety Disorder (GAD-7; Spitzer, Kroenke, Williams & Lowe, 2006) is a 7-item measure of anxiety symptoms with cutoff scores that indicate clinically significant levels of generalized anxiety (Spitzer et al., 2006).
Both questionnaires have the scale options (0 – “Not at all,” 1 – “Several days,” 2 – “More than half the days,”, 3 – “Nearly every day”).
```{r}
#| code-summary: "Time 1 dataset"
library(gt)
library(DT)
datatable(time1data)
```
::: panel-tabset
### Model 1: EBICglasso
Model 1 — The depression and anxiety symptom networks were estimated as Gaussian graphical models (GGM) separately at each wave using graphical LASSO regularization with EBIC.
The package is `bootnet::estimateNetwork` with "EBICglasso".
This is same as using `qgraph::EBICglasso()`.
The default setting of this function is converting input data into polychoric correlation matrix.
```{r}
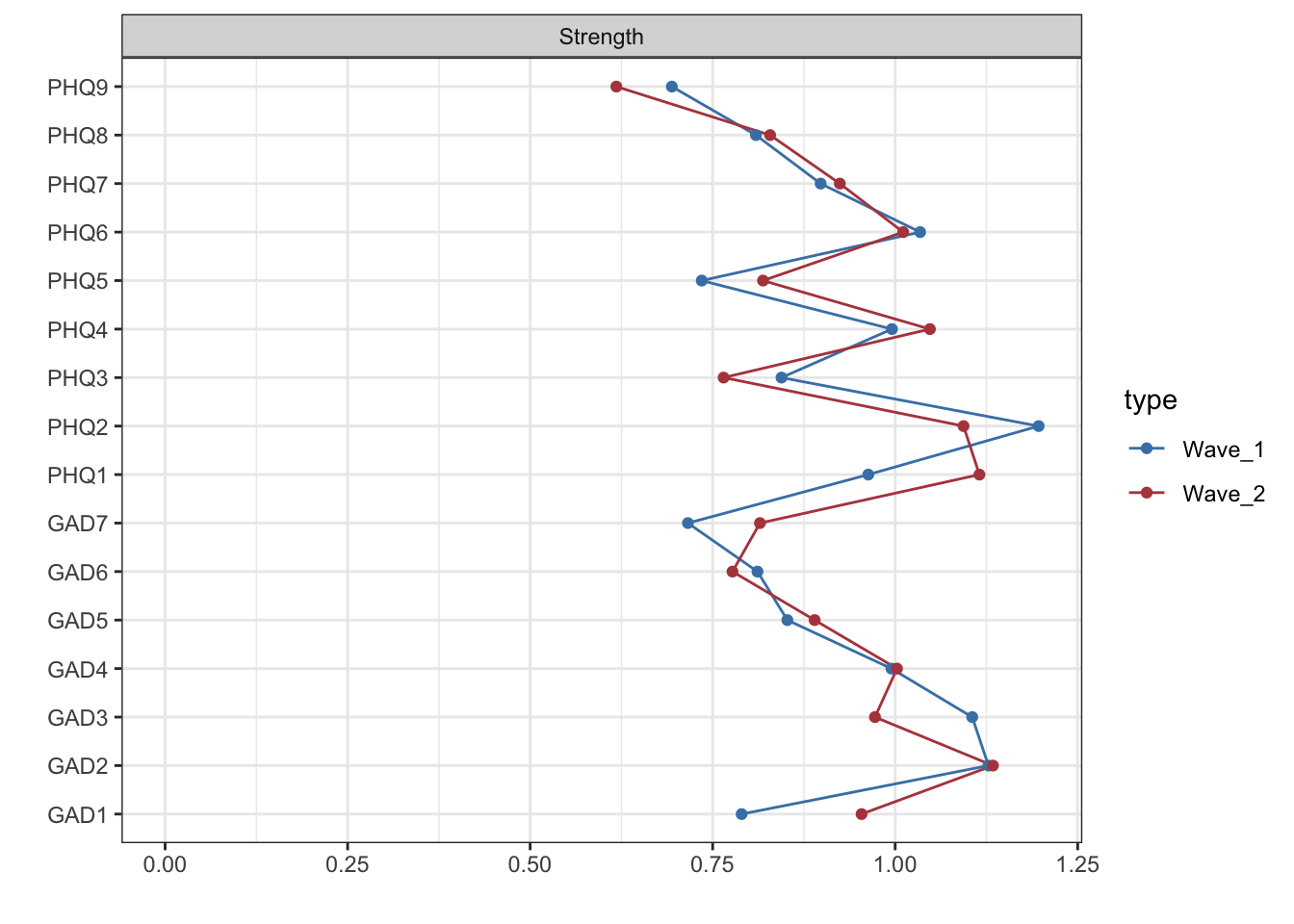
#| code-summary: "Visualize strength centrality for wave 1 and 2"
library(bootnet)
library(qgraph)
mycolors = c("#4682B4", "#B4464B", "#752021",
"#1B9E77", "#66A61E", "#D95F02", "#7570B3",
"#E7298A", "#E75981", "#B4AF46", "#B4726A")
##Estimate GGM networks
network1 <- estimateNetwork(time1data, default="EBICglasso")
network2 <- estimateNetwork(time2data, default="EBICglasso")
layout(t(1:1))
centw1w2 <- centralityPlot(list(Wave_1 = network1, Wave_2 = network2),
labels=labels_time1and2data, verbose = FALSE,
print = F)
centw1w2 + scale_color_manual(values = mycolors)
```
```{r}
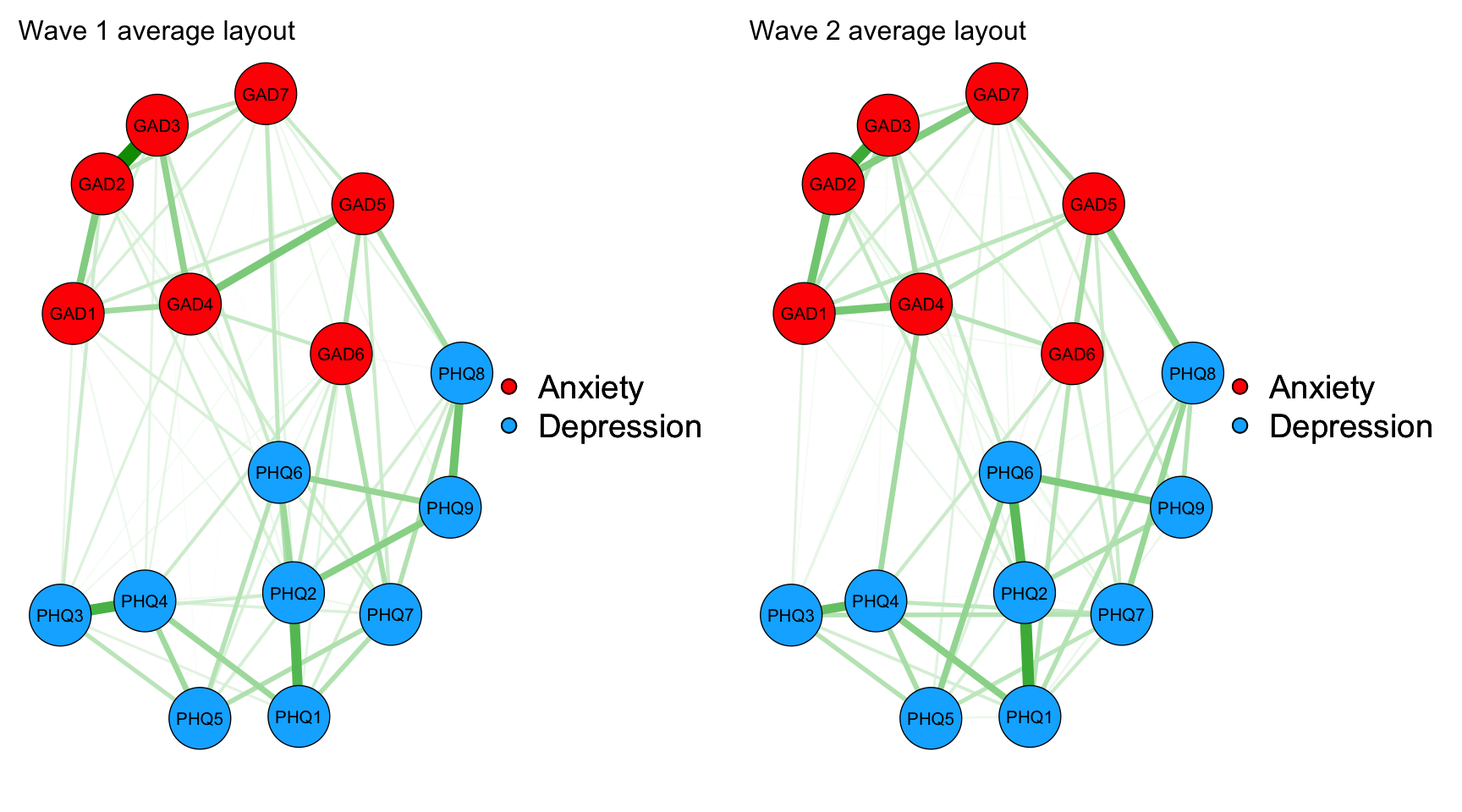
#| code-summary: "Visualize network structure for wave 1 and 2"
#| fig-width: 9
L <- averageLayout(network1, network2)
#Plot networks with average layout and edges comparable in weight
maxIsing<-max(c(network1$graph, network2$graph))
layout(t(1:2))
network1plot_average <- qgraph(network1$graph, layout=L, title="Wave 1 average layout", maximum=maxIsing, groups = groups_time1and2data)
network2plot_average <- qgraph(network2$graph, layout=L, title="Wave 2 average layout", maximum=maxIsing, groups = groups_time1and2data)
```
```{r network_characteristics}
#| code-summary: "Network characteristics for wave 1 and 2"
Networ_char_labes <- c(
NS_sum = "Global strength (sum absolute edge weights)",
NS_max = "Maximum strength",
NS_min = "Minimum strength",
Connectivity = "Connectivity (percentage of edges that are non-zero)",
Num_total_edges = "Total number of possible edges",
Num_nonzero_edges = "Number of non-zero edges",
Num_nonzero_edges = "Number of boostrapped non-zero edges",
Num_zero_edges = "Number of zero edges",
Num_positive_edges = "Number of positive edges",
Num_nonzero_edges = "Number of boostrapped positive edges",
Num_negative_edges = "Number of negative edges"
)
if (file.exists("tables/network_char_tbl.csv")){
network_char_tbl <- read.csv("tables/network_char_tbl.csv")
}else{
network_char_tbl <- data.frame(
`Network characteristics` = Networ_char_labes,
Wave1 = network_char(network1),
Wave2 = network_char(network2)
)
write.csv(network_char_tbl, file = "tables/network_char_tbl.csv", row.names = FALSE)
}
```
```{r}
#| echo: false
datatable(network_char_tbl)
```
```{r}
#| code-summary: "Most influential nodes for wave 1 and 2 based on node strength"
library(tidyverse)
centralityTable(list(Wave_1 = network1,
Wave_2 = network2)) |>
dplyr::filter(measure == "Strength") |>
group_by(type) |>
dplyr::filter(value == max(value)) |>
kableExtra::kable()
```
### Model 2: FIML with prune
```{r}
#| code-summary: "Esimate the network with psychonetrics"
library(psychonetrics)
network1_mod2 <- ggm(time1data, estimator = "FIML", standardize = "z") %>%
prune(alpha = .01, recursive = FALSE)
network2_mod2 <- ggm(time2data, estimator = "FIML", standardize = "z") %>%
prune(alpha = .01, recursive = FALSE)
network1_mod2_graph <- getmatrix(network1_mod2, "omega")
network2_mod2_graph <- getmatrix(network2_mod2, "omega")
```
```{r}
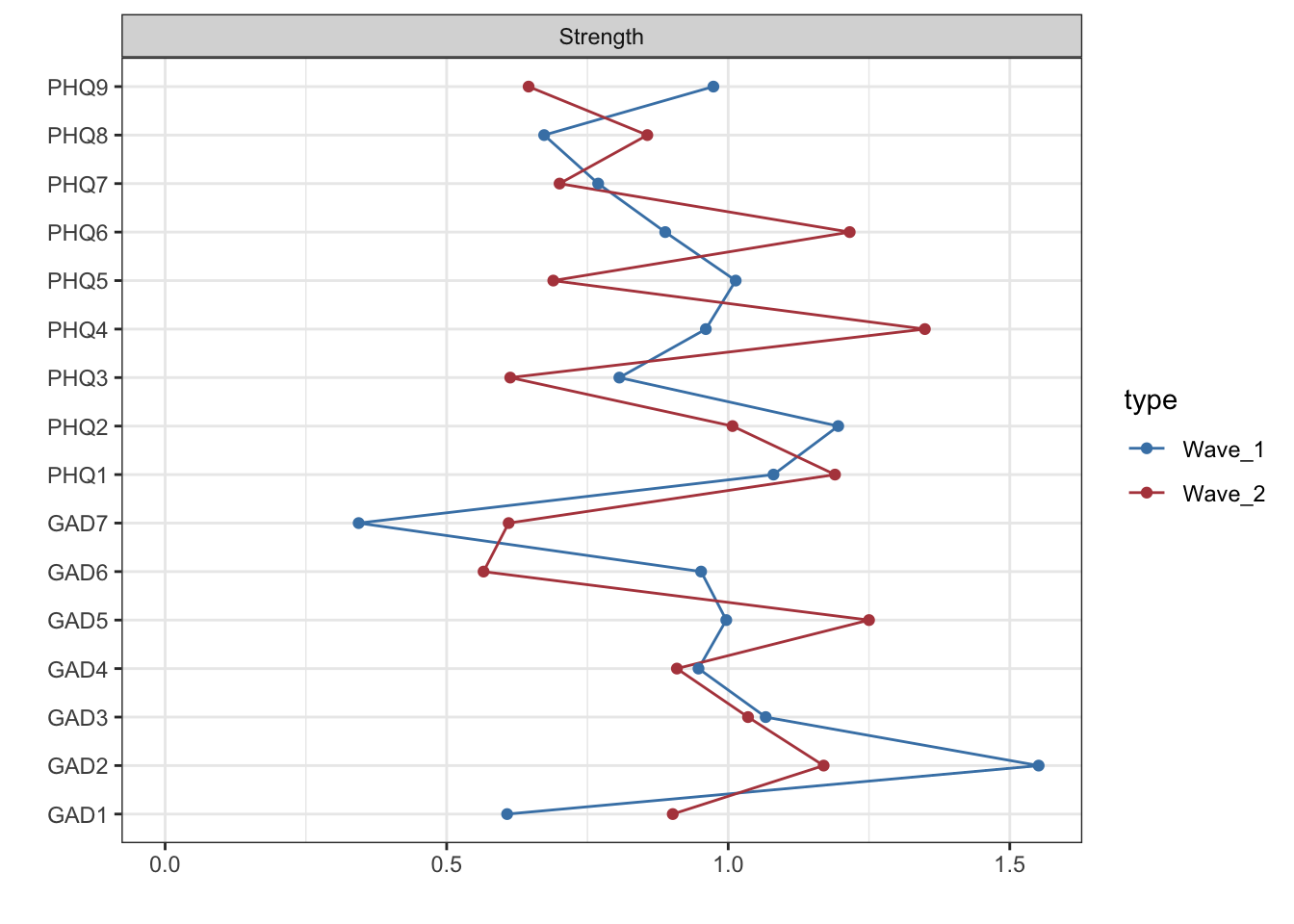
#| code-summary: "Visualize strength centrality for wave 1 and 2"
centw1w2_mod2 <- centralityPlot(list(Wave_1 = network1_mod2_graph, Wave_2 = network2_mod2_graph),
labels=labels_time1and2data, verbose = FALSE,
print = F)
centw1w2_mod2 + scale_color_manual(values = mycolors)
```
```{r}
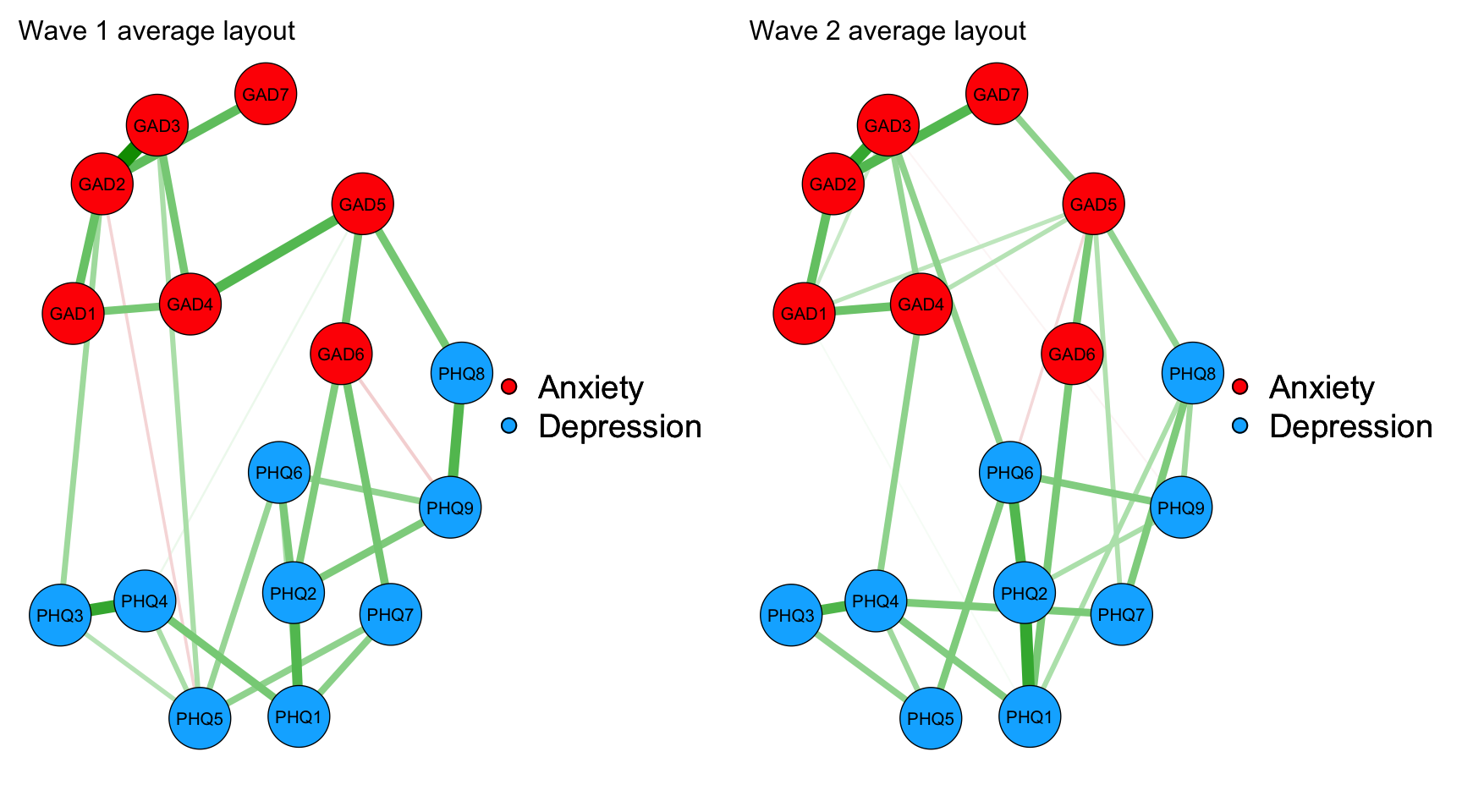
#| code-summary: "Visualize network structure for wave 1 and 2"
#| fig-width: 9
layout(t(1:2))
maxIsing2<-max(c(network1_mod2_graph, network2_mod2_graph))
network1_mod2_plot_average <- qgraph(network1_mod2_graph,
layout=L,
title="Wave 1 average layout",
labels=labels_time1and2data,
groups = groups_time1and2data,
maximum=maxIsing2)
network2_mod2_plot_average <- qgraph(network2_mod2_graph,
layout=L,
title="Wave 2 average layout",
labels=labels_time1and2data,
groups = groups_time1and2data,
maximum=maxIsing2)
```
:::
## Results
## Discussion