---
title: "Lecture 01: Basics of R"
subtitle: "Getting Started"
author: "Jihong Zhang*, Ph.D"
institute: |
Educational Statistics and Research Methods (ESRM) Program*
University of Arkansas
date: "2024-10-09"
date-modified: "2024-10-11"
sidebar: id-lec6990v
execute:
echo: true
warning: false
eval: false
output-location: default
code-annotations: below
format:
html:
code-tools: true
code-line-numbers: false
code-fold: false
number-offset: 0
anchor-sections: true
number-sections: false
uark-revealjs:
scrollable: true
chalkboard: true
embed-resources: false
code-fold: false
number-sections: false
footer: "ESRM 64503 - Lecture 01: Introduction to R"
slide-number: c/t
tbl-colwidths: auto
output-file: slides-index.html
filters:
- quarto
- line-highlight
---
## Today's Class
1. Why using R?
1. Brief history of R
2. Main features of R
2. Installation of R
3. How to use RStudio
# Why R?
## Brief History
- **1975-1976**: S (Book: [*A Brief History of S*](https://sas.uwaterloo.ca/~rwoldfor/software/R-code/historyOfS.pdf)) grew up in the statistics research departments (John Chambers and others) at Bell Laboratories
- To bring interactive computing to bear on statistics and data analysis problem
- **1993**: Prof. [Ross Ihaka](https://en.wikipedia.org/wiki/Ross_Ihaka "Ross Ihaka") and [Robert Gentleman](https://en.wikipedia.org/wiki/Robert_Gentleman_(statistician)) from University of Auckland posted first binary file of R to teach introductory statistics
- **1995**: Martin Mächler made an important contribution by convincing Ross and Robert to use the [GNU General Public License](http://www.gnu.org/licenses/gpl-2.0.html) to make R free software
- **1997**: The [Comprehensive R Archive Network](https://en.wikipedia.org/wiki/R_package#Comprehensive_R_Archive_Network_(CRAN) "R package") (**CRAN**) was founded by Kurt Hornik and [Friedrich Leisch](https://en.wikipedia.org/wiki/Friedrich_Leisch "Friedrich Leisch") to host R's [source code](https://en.wikipedia.org/wiki/Source_code "Source code"), executable files, documentation, and user-created packages
- **2000**: the first official 1.0 version of R was released
- **2024**: R ver. 4.2.1
## Example of S Language
```{r}
#| error: true
#| output-location: column
#| eval: true
X = 1:5 # A vector of numbers from 1 to 5
X[c(TRUE, TRUE, TRUE, FALSE, FALSE)]
X[1:3]
X[-1:3]
X[-(1:3)]
X[NULL]
X[NA]
X[]
```
## Main Feature of R
1. it was developed by statisticians as an interactive environment for data analysis rather than C or Java that created by software development.
2. The interactivity of R is an indispensable feature in data science
3. However, like in other programming languages, you can save your work in R as scripts that can be easily executed at any moment.
4. If you are an expert programmer, you should not expect R to follow the conventions you are used to since you will be disappointed.
## Attractive Features of R
- R is free and open source.
- It runs on all major platforms: Windows, MacOS, UNIX/Linux.
- Scripts and data objects can be shared seamlessly across platforms.
- There is a large, growing, and active community of R users and, as a result, there are numerous resources for learning and asking questions.
- [stackoverflow](https://stats.stackexchange.com/questions/138/free-resources-for-learning-r)
- [r-project.com](https://www.r-project.org/help.html)
- It is easy for others to contribute add-ons which enables developers to share software implementations of new data science methodologies. This gives R users early access to the latest methods and to tools which are developed for a wide variety of disciplines, including ecology, molecular biology, social sciences, and geography, just to name a few example
# Get started
## R console
- One way of using R is to simply start R console on your computer (PC).
- In Mac, after installing R, simply type in "R" in terminal to get started
::: panel-tabset
## Windows
{fig-align="center"}
## Mac/Linux
{fig-align="center"}
:::
As a quick example, try using the console to calculate a 15% tip on a meal that cost \$19.71:
```{r}
0.15 * 19.71
```
# Rstudio (now called Posit)
## Installation of R and RStudio
- You can download and install **R base** via r-project.org (currently R-4.4.1)
- for [Linux](https://cloud.r-project.org/bin/linux/)
- for [Windows](https://cloud.r-project.org/bin/windows/)
- for [MacOS](https://cloud.r-project.org/bin/macosx/)
- Then, after the installation of R, you can download RStudio via posit.co (currently)
- for [Windows](https://download1.rstudio.org/electron/windows/RStudio-2024.04.2-764.exe)
- for [MacOS](https://download1.rstudio.org/electron/macos/RStudio-2024.04.2-764.dmg)
- After installation of R and RStudio, you can open up the RStudio to start your R programming.
- however, your R only has the **base** package
- To enhance its utility, most users will install **R packages** for certain purposes
## R packages
- R packages are uploaded to some platforms (i.e., CRAN or Github) by researchers or companies
- Those R packages typically have their version numbers. Some functions may be available for some version (like Ver. 1.1) but not be available in other versions.
- Do not upgrade your packages if you code is running well
- R users are free to download and use those R packages
- To download certain package, you should know package name
- For example, if you want to download the latest version of `tidyverse` package, you can type in following command in **the console panel** of Rstudio
```{r}
#| eval: false
#| echo: true
#| code-fold: false
install.packages("tidyverse")
```
- Or if you want to install the older version of package
```{r}
#| eval: false
#| echo: true
#| code-fold: false
require(devtools)
install_version("tidyverse", version = "1.3.0", repos = "http://cran.us.r-project.org")
```
## More about R packages
- **CRAN** (Comprehensive R Archive Network) is a network of servers around the world that store identical, up-to-date, versions of code and documentation for R.
- It contains most stable version of packages.
- Most of time, we download package from CRAN
- **Github** is for the fast development for R packages
- It contains the up-to-date version of R which may potentially be unstable
- You can download the package from Github using `pak` package
```{r}
#| eval: false
#| echo: true
#| code-fold: false
pak::pak("tidyverse/ggplot2")
```
- You can update the package and its dependencies
```{r}
#| eval: false
#| echo: true
#| code-fold: false
pak::pkg_install("ggplot2", upgrade = TRUE)
```
## R functions
- To operate certain tasks, you need to use functions contained in R packages
- There are two ways of using R functions
- **Direct way**: you don't have to load your package first
- 
------------------------------------------------------------------------
- **Use-after-load way**: Package is loaded in your session before you can call the function name without specifying the package name
```{r}
#| eval: true
#| echo: true
#| code-fold: false
library("ggplot2")
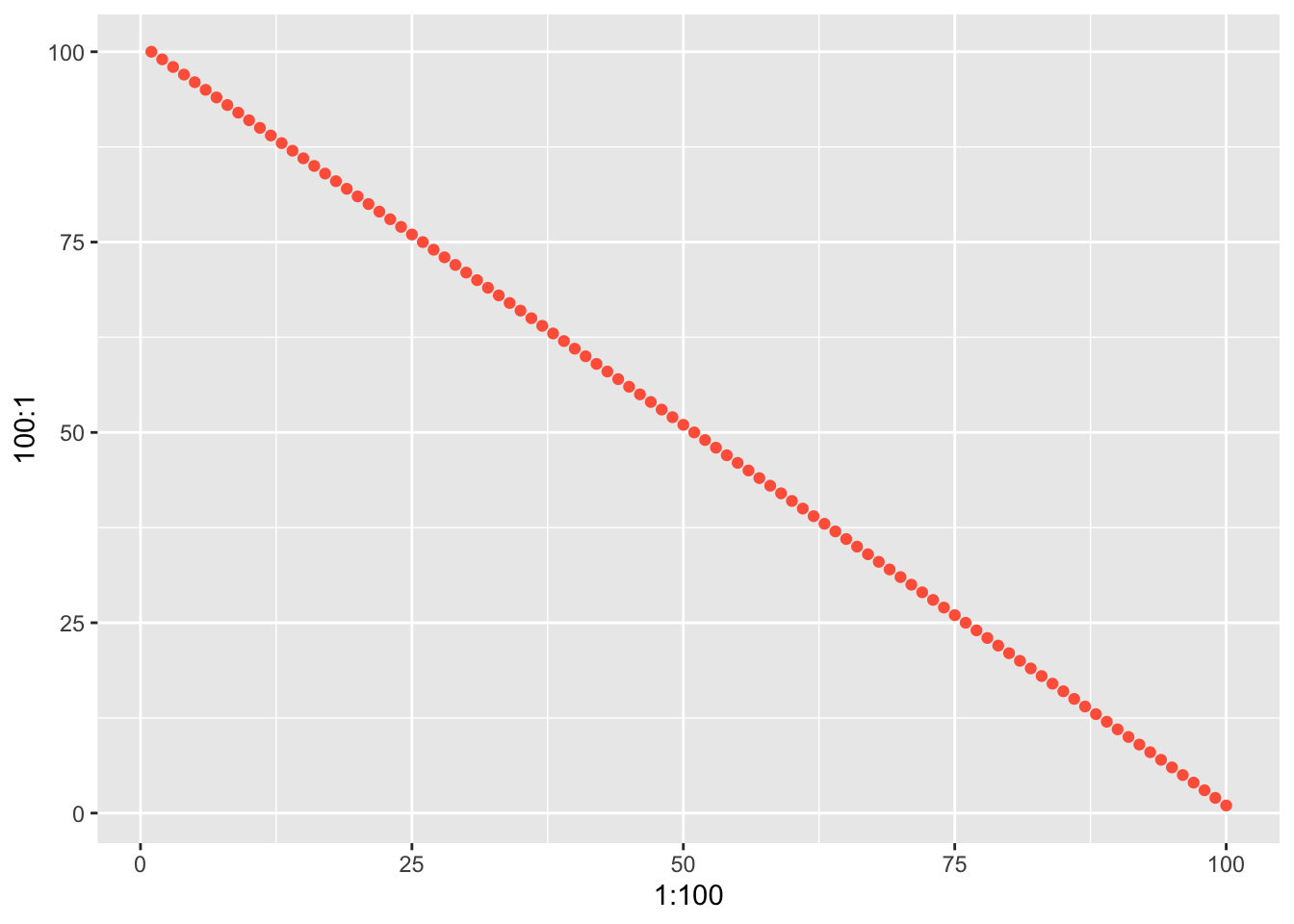
ggplot() +
geom_point(aes(x = 1:100, y = 100:1), color = "tomato")
```
## R functions (Cont.)
- How do you know you already load the package or not
- You can use `sessionInfo` function
```{r}
#| echo: true
#| code-fold: false
sessionInfo()
```
- It outputs multiple info:
- R version, Operations System, Matrix operation package, Locale
- **Attached packages** (you can call the functions of those package)
- **Loaded package via a namespace (and not attached)**, which you cannot call functions and need to `library` or `require` them
## Execute R code
- After you finish R script, you have **multiple ways of executing the code and output the results on Consol**e:
- **Method 1**: you can click `Run` button in the top right-head of **Rstudio**
- **Method 2**: you can select certain code and press `Ctrl + Enter` (Win) or `Command + Return` (Mac)
- **Method 3**: you can `Rscript [FILENAME].r` to run the whole script
- **Method 4**: you can using R notebook to interactively run R code
+----------------------------+-------------------+-----------------------------+
| | Script file is .R | Script file is .rmd or .qmd |
+============================+===================+=============================+
| **Run the whole script** | - Method 1 | - Method 4 |
| | - Method 3 | |
+----------------------------+-------------------+-----------------------------+
| **Run the partial script** | - Method 2 | - Method 4 |
+----------------------------+-------------------+-----------------------------+
: {.striped}
##
## Introduce Rstudio
- RStudio will be our launching pad for data science projects. It not only provides an editor for us to create and edit our scripts but also provides many other useful tools.
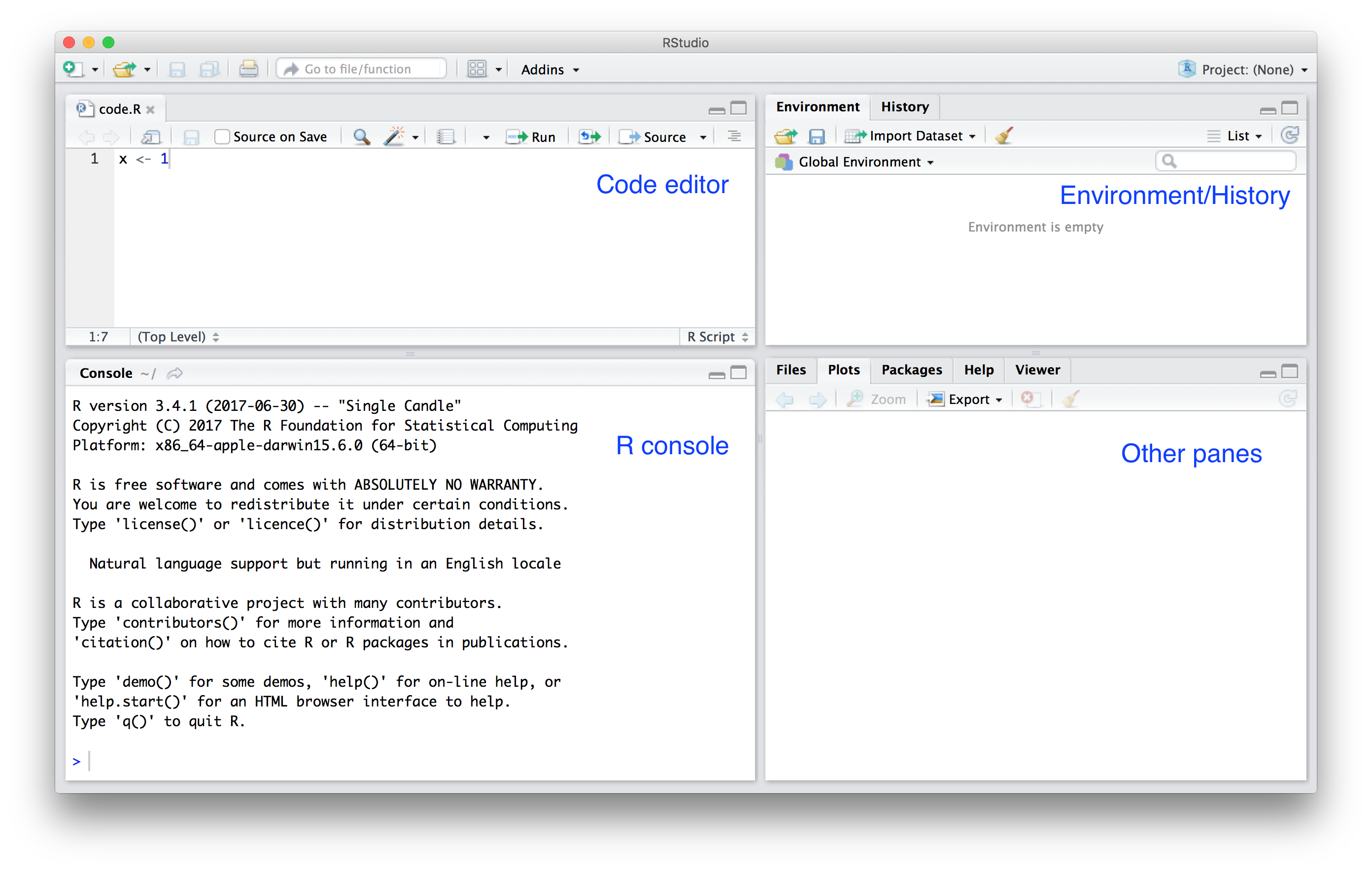
- When you start RStudio for the first time, you will see three panes:
- The left pane shows the Code editor (will show when you create a new file) and R console.
- On the right, the top pane includes tabs such as *Environment* and *History*, while the bottom pane shows five tabs: *File*, *Plots*, *Packages*, *Help*, and *Viewer* .
- To start a new script in Code editor, you can click on `File` \> `New File` \> `R Script`.
::: panel-tabset
## Screenshot of Rstudio
{fig-align="center"}
## Start a new script
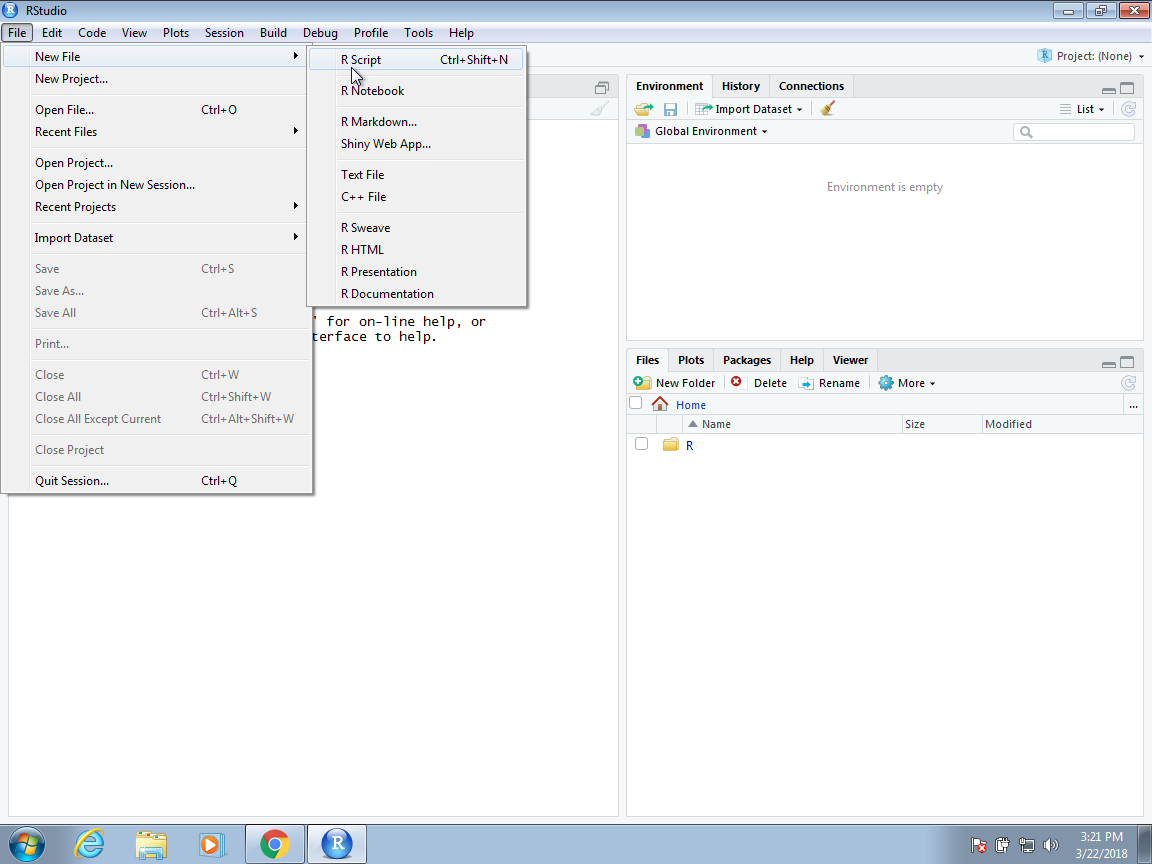{fig-align="center"}
## Start writing your script
{fig-align="center"}
:::
## Key Binding
- For the efficient coding, **we highly recommend that you memorize key bindings for the operations you use most**.
- RStudio provides a useful cheat sheet with the most widely used commands
- To open the cheat sheet, `Help` \> `Cheat Sheets` \> `Rstudio IDE Cheat Sheets`
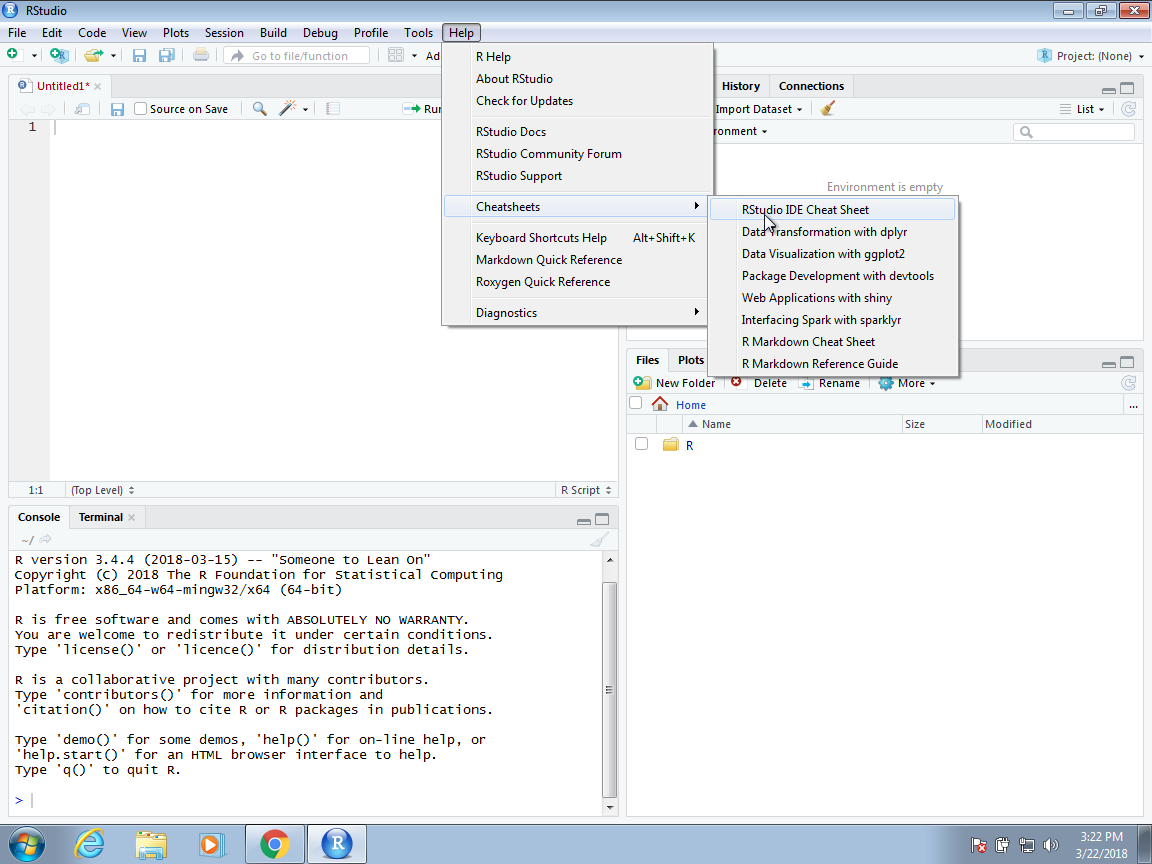{fig-align="center"}
## Global Option
- You can change the look and functionality of RStudio quite a bit.
- To change the global options you click on *`Tools`* then *`Global Options`…*.
- As an example we show how to make a change that we **highly recommend**:
- `General` \> `Basic` \> `Workspace`: Change the [*Save workspace to .RData*]{.underline} *on exit* to [*Never*]{.underline} .
- `General` \> `Basic` \> `Workspace`: Uncheck the [*Restore .RData into workspace at startup*]{.underline} to [*Never*]{.underline}
- `Code` \> `Editing`: check [*use the native piper operator, \|\>*]{.underline}
::: callout-note
## .RData file
- By default, when you exit R saves all the objects you have created into a file called .RData.
- This is done so that when you restart the session in the same folder, it will load these objects.
- We find that this causes confusion especially when we share code with colleagues and assume they have this .RData file.
:::
## Installing R Packages: from CRAN
- For example, to install the **`dslabs`** package, you would type the following in your console:
```{r}
install.packages("dslabs") # DON'T FORGET DOUBLE QUOTE
```
- We can then load the package into our R sessions using the `library` function in your Rscript file:
```{r}
#| eval: true
library("dslabs")
head(admissions)
```
- As you go through this class, you will see that we load packages without installing them. This is because once you install a package, it remains installed and only needs to be loaded with `library`.
- We can install more than one package at once by feeding a character vector to this function:
```{r}
install.packages(c("dplyr", "dslabs"))
```
- You can see all the packages you have installed using the following function:
```{r}
installed.packages()
```
## Installing R Packages: from GitHub
- You can also install user-built package from GitHub
- I built a package for this course: [link](https://github.com/JihongZ/ESRM6990V)
```{r}
#| eval: false
install.packages("remotes") # install one package called "remotes"
library("remotes") # load the package into your R session
install_github(repo = "JihongZ/ESRM6990V") # install one GitHub package from my GitHub repository
library(ESRM6990V) # load the package into your R session
jihong(details = TRUE) # call one function called "jihong" from the package
```
### Let's Practice
1. [Finish Exercise 1](ESRM6990V_Example01.qmd)
# R Package Structure
## Basic Information
1. **What**: An R package is a structured collection of R functions, data, and compiled code that is bundled together according to a specific format.
- They can be thought of as libraries or modules in other programming languages.
2. **Why**: R Packages are designed to add functionality to R, allowing users to perform specific tasks or analyses that are not covered by the basic installation of R.
3. **How:** You can install/uninstall, create, load, and use R packages.
- If you want to build or publish your own package, the Comprehensive R Archive Network (CRAN), Bioconductor, and GitHub are popular repositories where R packages are commonly published and maintained.
## What R package include
- **Functions**: A set of R functions that perform specific tasks, which are not available in the default R environment.
- **Data**: Some packages include datasets that are useful for demonstrating functions within the package or for use in specific types of analysis.
- **Documentation**: Every package comes with documentation that explains how the functions work, the data included (if any), and examples of how to use the package. This is often accessible via R help pages.
- **Vignettes**: Many packages include vignettes, which are long-form documentation that shows how to use the package functions in a more detailed and contextual way, often in the form of tutorials.
- **Namespace**: A namespace file that manages how functions from the package are imported and exported, helping avoid naming conflicts between different packages.
- **Meta-information**: A DESCRIPTION file containing metadata about the package, such as its name, version, dependencies (other packages it requires to function), author, and license information.
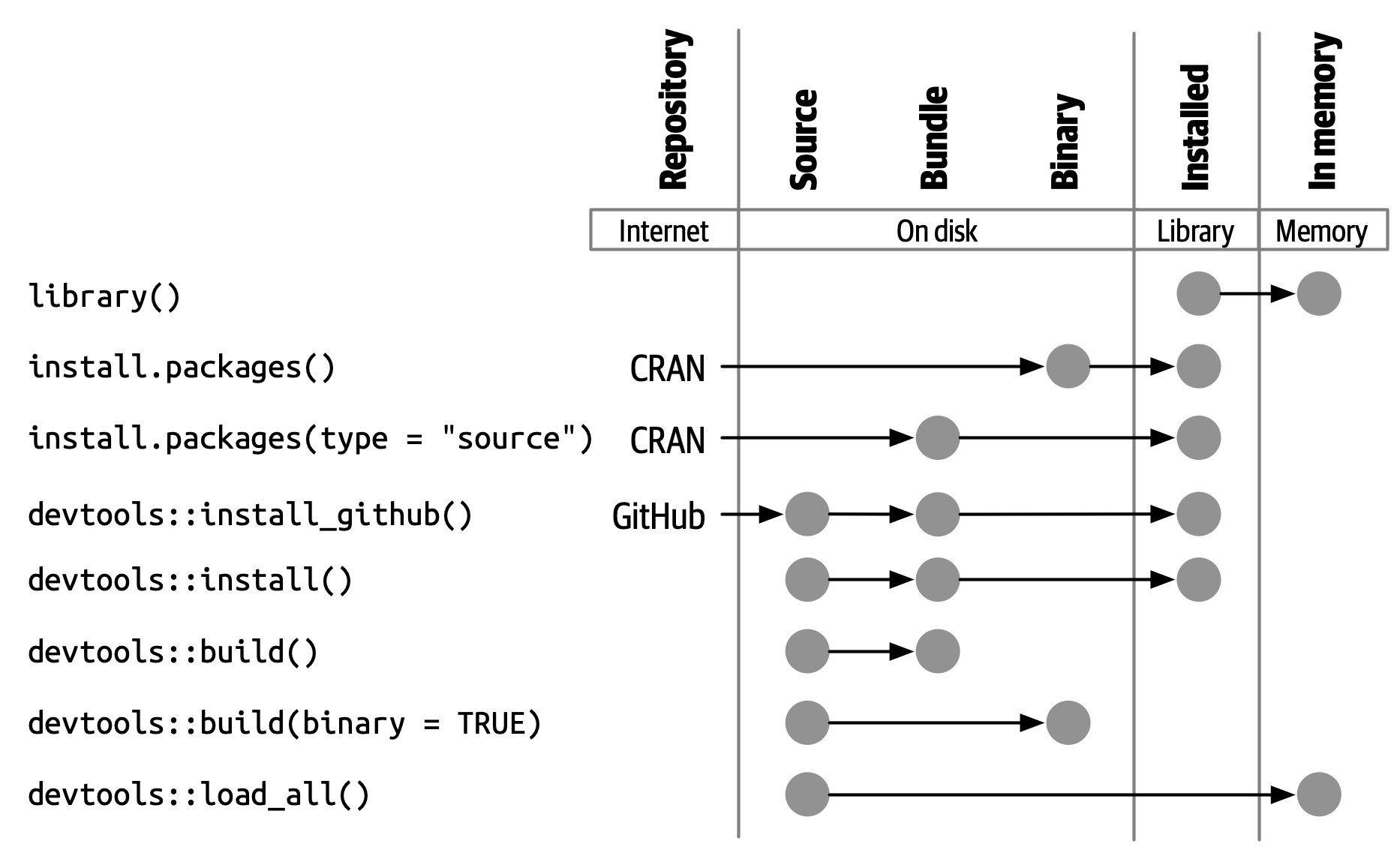
## R Package states
1. When you create or modify a package, you work on its “source code” or “source files”. You interact with the in-development package in its **source** form.
2. To better understand package, we need to know the five states of R package:
1. source
2. bundled
3. binary
4. installed
5. in-memory
3. We already know two functions:
1. `install.packages()` can move a package from **source/bundled/binary** into **installed** state.
2. `library` can load a package from installed state into memory (**in-memory** state)
4. What are source/bundled/binary states then? Why they differ?
## Source package
1. A source package is just a directory of files with specific structure including:
1. **DESCRIPTION** file
2. `R/` folder containing all `.r` files
2. Many R packages on GitHub are in source state
1. `networkscore`: <https://github.com/JihongZ/networkscore>
2. `esrm64503`: <https://github.com/JihongZ/ESRM64503>
3. You may also find some `tar.gz` file on packages' CRAN landing page via the “Package source” field (this is the bundled state of the package). Decompressing the `tar.gz` file will have the source directory including `R/` and `DESCRIPTION`
1. forcats: <https://cran.r-project.org/web/packages/forcats/index.html>
2. You can depress using commands in terminal like:
``` bash
tar xvf forcats_0.4.0.tar.gz
```
## Bundled package
1. A bundled package is a package that's been compressed into a single file (this process is called `build` the package). Bundled state is a compressed form of package with only single file.
2. By convention, package bundles in R use the extension `.tar.gz` and are sometimes referred to as "source tarballs". In computer science, it is called gzipped tar file format.
3. A "source tarballs" file is not simply compressed file of source directory. When build source directory into bundled (.tar.gz), a few diagnostic checks and cleanups are performed. See more details [here](https://cran.r-project.org/doc/manuals/R-exts.html#Building-package-tarballs).
## Binary package
1. If you want to distribute your package to an R user who doesn’t have package development tools, you’ll need to provide a **binary** package. The main distributor of **binary** package is CRAN.
2. Like a package bundle, a binary package is a single file. Unlike a bundled package, a binary package is platform specific and there are two basic flavors: Windows and macOS.
3. CRAN packages are usually available in binary form:
- forcats for macOS: forcats_0.4.0.tgz
- readxl for Windows: readxl_1.3.1.zip
4. This is, indeed, part of what’s usually going on behind the scenes when you call `install.packages()`.
5. Uncompressing binary file will give you totally difference file structure than source/bundled package.
- There are no .R files in the R/ directory - instead there are three files that store the parsed functions in an efficient file format.
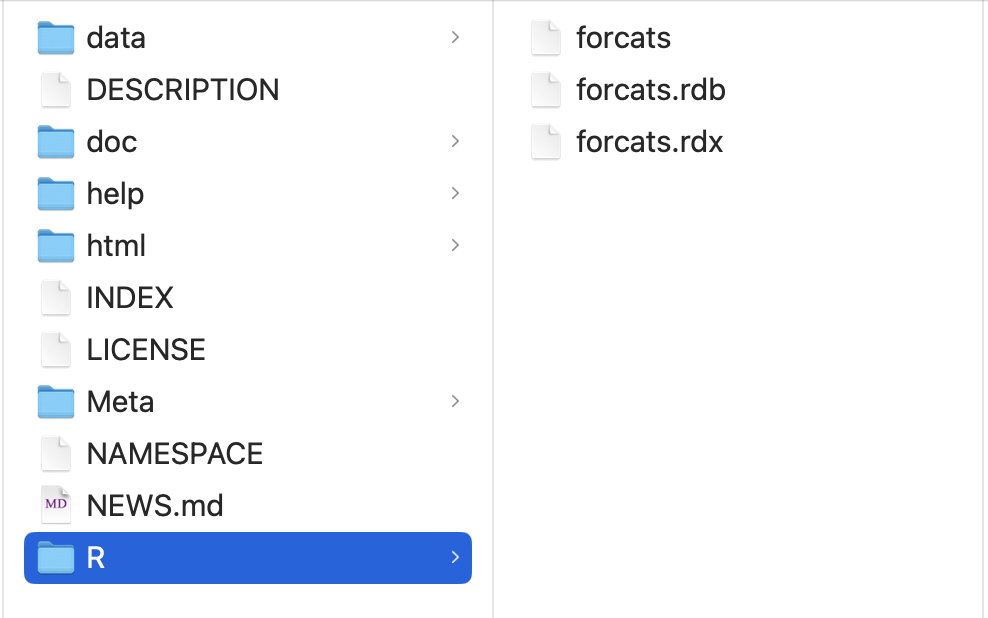
## Installed package
1. An installed package is a binary package that's been decompressed into a package library
2. In practice, you don't need to care about stats if you install popular package, unless you have issues installing R package via `install.packages()` or you install in-development packages .
## In-memory package
1. When we use `library()` function, we load installed package into the memory of R.
2. This is the last step of using the package in our R task.
3. When you call `library(somepackage)`, R looks through the current libraries for an installed package named “somepackage” and, if successful, it makes somepackage available for use.
# Next Week
## Preparation: Make Contribute to Github R Package
1. Make sure you have set up a [GitHub account](https://github.com/)
2. Make sure you download the [GitHub Desktop](https://desktop.github.com/download/)