Click here to see R code
dog_means <- c(19.25, 20.75, 22.25, 17.50, 18.75, 19.50 )
sum(4 * (dog_means - 19.67)^2)[1] 54.3336Experimental Design in Education
Jihong Zhang*, Ph.D
Educational Statistics and Research Methods (ESRM) Program*
University of Arkansas
April 6, 2025
April 10, 2025
Question: How about the research scenario when more than two independent variables?
Note. All ANOVA-related designs talked about in this class can handle only one dependent variable; if you want to deal with more than 2 dependent variable, we may consider other research designs…
To conduct the hypothesis test, we follow:
Before conducting the hypothesis test, we check:
Independence of the observations:
| Tutoring Type | Time 1 (1st week out) | Time 2 (2nd week out) | Time 3 (3rd week out) |
|---|---|---|---|
| None | Sally Joe Montes |
Sally Joe Montes |
Sally Joe Montes |
| Once/week | Omar Fred Jenny |
Omar Fred Jenny |
Omar Fred Jenny |
| Daily | Chi Lena Caroline |
Chi Lena Caroline |
Chi Lena Caroline |
Two types of repeated measures are common:
Cross-over Design: alternatively, experiments can involve administering all treatment levels (in a sequence) to each experimental unit.
| Period 1 | Period 2 | |
|---|---|---|
| Sequence AB | A | B |
| Sequence BA | B | A |
How many potential sequences are for J x J design?
\prod (J, J-1, \cdots,1)
Two types of repeated measures are common:
| Period 1 | Period 2 | Period 3 | |
|---|---|---|---|
| Sequence ABC | A | B | C |
| Sequence BCA | B | C | A |
| Sequence CAB | C | A | B |
| Sequence ACB | A | C | B |
| Sequence BAC | B | A | C |
| Sequence CBA | C | B | A |
Carryover effects:
In a cross-over design, carryover effects refer to the residual effects of a treatment that persist and influence responses in subsequent treatment periods.
Since each participant receives multiple treatments in sequence, the outcome measured in a later period may be affected not only by the current treatment but also by the lingering influence of prior treatments.
✔ Example:
Discussion: Can you think about more carryover effect in your research area?
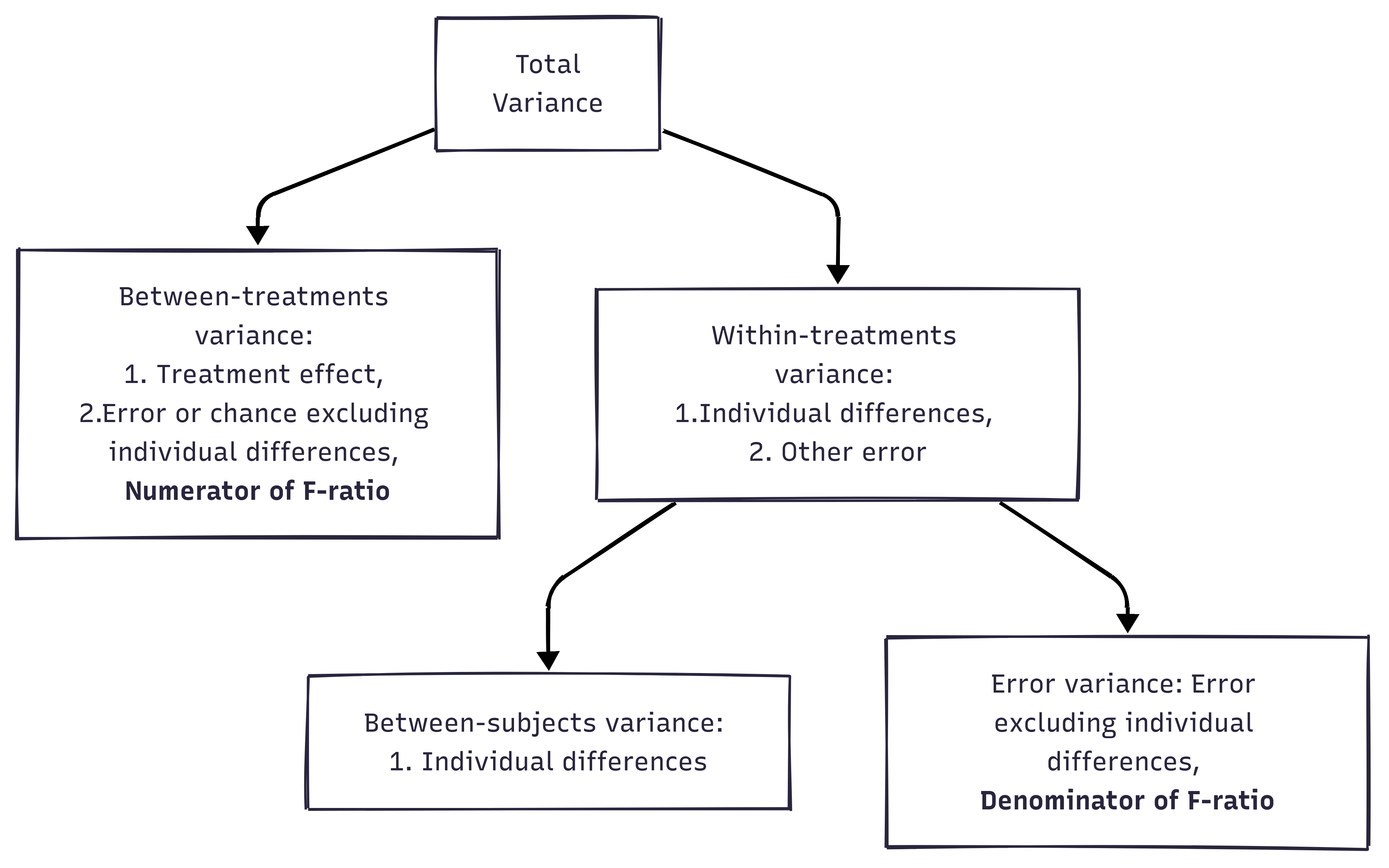
In RM ANOVA design, within-group variability (SSw) is used as the error variability (SSError).
The F-statistic is calculated as the ratio of MSModel to MSError:
Independent ANOVA:
F = \frac{MS_b}{MS_w} = \frac{MS_b}{MS_{error}}
“Why subjects differ within the same group (e.g., treatment/control groups)” can be explained in RM ANOVA!
The advantage of a repeated measures ANOVA:
A repeated measures ANOVA calculates an F-statistic in a similar way:
 Generated by https://www.mermaidchart.com/app/projects
Generated by https://www.mermaidchart.com/app/projects
Mathematically, we are partitioning the variability attributable to the differences between groups (SSconditions) and variability within groups (SSw) exactly as we do in a between-subjects (independent) ANOVA.
With a repeated measures ANOVA, we are using the same subjects in each group, so we can remove the variability due to the individual differences between subjects, referred to as SSsubjects, from the within-groups variability (SSw).
We simply treat each subject as a “block.”
The ability to subtract SSsubjects will leave us with a smaller SSerror term:
Independent ANOVA: SS_{error} = SS_{w}
Repeated Measures ANOVA: SS_{error} = SS_{w} - SS_{subjects}
| Dog ID | time1 | time2 | time3 | time4 | Dog mean |
|---|---|---|---|---|---|
| 1 | 10 | 16 | 25 | 26 | 19.25 |
| 2 | 12 | 19 | 27 | 25 | 20.75 |
| 3 | 13 | 20 | 28 | 28 | 22.25 |
| 4 | 12 | 18 | 25 | 15 | 17.50 |
| 5 | 11 | 20 | 26 | 18 | 18.75 |
| 6 | 10 | 22 | 27 | 19 | 19.50 |
| Time mean | 11.33 | 19.17 | 26.33 | 21.83 | Grand mean = 19.67 |
| Source | Sub-Component | SS | df | MS | F |
|---|---|---|---|---|---|
| Between-time | |||||
| Within-time (I) | Between-Subjects | ||||
| Within-time (II) | Within-Subjects / Error |
Note. Under the repeated measures ANOVA, we can decompose the total variability:
Total variability of DV = “between-time” variability + “within-time” variability
Between-subjects are useful because we can use it to explain the treatment effects / gender effects (anything relevant to why people different from each other).
We calculate mean square for between-time as:
MS_{btime} = \frac{SS_{btime}}{df_{btime}}
Where:
SS_{btime} = \sum n_{subjects}(\bar{Y}_{time} - \bar{Y}_{grand})^2 = 6(11.33 - 19.67)^2 + \cdots + 6(21.83 - 19.67)^2 = 712.961
df_{btime} = \text{Number of time points} - 1 = 4 - 1 = 3
Thus,
MS_{btime} = \frac{712.96}{3} = 237.65
| Dog ID | time1 | time2 | time3 | time4 | Dog mean |
|---|---|---|---|---|---|
| 1 | 10 | 16 | 25 | 26 | 19.25 |
| 2 | 12 | 19 | 27 | 25 | 20.75 |
| 3 | 13 | 20 | 28 | 28 | 22.25 |
| 4 | 12 | 18 | 25 | 15 | 17.50 |
| 5 | 11 | 20 | 26 | 18 | 18.75 |
| 6 | 10 | 22 | 27 | 19 | 19.50 |
| Time means | 11.33 | 19.17 | 26.33 | 21.83 | Grand Mean = 19.67 |
| Source | SS | df | MS | F |
|---|---|---|---|---|
| Between-time | 712.96 | 3 | 237.65 | |
| Within-time | ||||
| Between-Subjects | ||||
| Error |
We calculate mean square for between-subjects (between-dog) as:
MS_{bsubjects} = \frac{SS_{bsubjects}}{df_{bsubjects}}
Where:
SS_{bsubjects} = \sum n_{times}(\bar{Y}_i - \bar{Y}_{grand})^2 = 4*(19.25 - 19.67)^2 + \cdots + 4*(19.50 - 19.67)^2 = 54.33
[1] 54.3336df_{bsubject} = N_{dogs} - 1 = 6 - 1 = 5
Thus,
MS_{bsubjects} = \frac{54.33}{5} = 10.866
| Dog ID | time1 | time2 | time3 | time4 | Dog mean |
|---|---|---|---|---|---|
| 1 | 10 | 16 | 25 | 26 | 19.25 |
| 2 | 12 | 19 | 27 | 25 | 20.75 |
| 3 | 13 | 20 | 28 | 28 | 22.25 |
| 4 | 12 | 18 | 25 | 15 | 17.50 |
| 5 | 11 | 20 | 26 | 18 | 18.75 |
| 6 | 10 | 22 | 27 | 19 | 19.50 |
| Time means | 11.33 | 19.17 | 26.33 | 21.83 | Grand Mean = 19.67 |
| Source | SS | df | MS | F |
|---|---|---|---|---|
| Between-time | 712.96 | 3 | 237.65 | |
| Within-time | ||||
| Between-Subjects | 54.33 | 5 | 10.87 | |
| Error |
We calculate the mean square for within-time as:
MS_{wtime} = \frac{SS_{wtime}}{df_{wtime}}
Where:
SS_{wtime} = \sum_{i,time}(y_{i,time} - \bar{Y}_{time})^2 = (10 - 11.33)^2 + \cdots + (19 - 21.83)^2 = 170.33
df_{wtime} = (6 - 1) + (6 - 1) + (6 - 1) + (6 - 1) = 20
Thus,
MS_{wtime} = \frac{170.33}{20} = 8.5165
| Dog ID | time1 | time2 | time3 | time4 | Dog mean |
|---|---|---|---|---|---|
| 1 | 10 | 16 | 25 | 26 | 19.25 |
| 2 | 12 | 19 | 27 | 25 | 20.75 |
| 3 | 13 | 20 | 28 | 28 | 22.25 |
| 4 | 12 | 18 | 25 | 15 | 17.50 |
| 5 | 11 | 20 | 26 | 18 | 18.75 |
| 6 | 10 | 22 | 27 | 19 | 19.50 |
| Time means | 11.33 | 19.17 | 26.33 | 21.83 | Grand Mean = 19.67 |
| Source | SS | df | MS | F |
|---|---|---|---|---|
| Between-time | 712.96 | 3 | 237.65 | |
| Within-time | 170.33 | 20 | 8.52 | |
| Between-Subjects | 54.33 | 5 | 10.87 | |
| Error |
We calculate the mean square for error as:
MS_{error} = \frac{SS_{error}}{df_{error}}
Where:
SS_{error} = SS_{wtime} - SS_{bsubjects} = 170.33 - 54.33 = 116
df_{error} = df_{wtime} - df_{bsubjects} = 20 - 5 = 15
Thus:
MS_{error} = \frac{116}{15} = 7.333
| Dog ID | time1 | time2 | time3 | time4 | Dog mean |
|---|---|---|---|---|---|
| 1 | 10 | 16 | 25 | 26 | 19.25 |
| 2 | 12 | 19 | 27 | 25 | 20.75 |
| 3 | 13 | 20 | 28 | 28 | 22.25 |
| 4 | 12 | 18 | 25 | 15 | 17.50 |
| 5 | 11 | 20 | 26 | 18 | 18.75 |
| 6 | 10 | 22 | 27 | 19 | 19.50 |
| Time means | 11.33 | 19.17 | 26.33 | 21.83 | Grand Mean = 19.67 |
| Source | SS | df | MS | F |
|---|---|---|---|---|
| Between-time | 712.96 | 3 | 237.65 | |
| Within-time | 170.33 | 20 | 8.52 | |
| Between-Subjects | 54.33 | 5 | 10.87 | |
| Error | 116.00 | 15 | 7.33 |
Research Question:
Does histamine concentration change over time?
(That is, does mean histamine concentration differ across the four time points?)
(Histamine is a chemical your immune system releases.)
Null Hypothesis:
H_0: \mu_{time1} = \mu_{time2} = \mu_{time3} = \mu_{time4}
ANOVA Table
| Source | SS | df | MS | F |
|---|---|---|---|---|
| Between-time | 712.96 | 3 | 237.65 | 237.65 / 7.33 = 30.73 |
| Within-time | 170.33 | 20 | 8.52 | 8.52 / 7.33 = 1.16 |
| Between-Subjects | 54.33 | 5 | 10.87 | 10.87 / 7.33 = 1.48 |
| Error | 116.00 | 15 | 7.33 |
Conclusion
Under \alpha = 0.05, the critical value for df_{time} = 3 and df_{error} = 15 from the F-table is:
F_{crit} = 3.29
Since the observed F-value for the time effect is 30.73, which exceeds the critical value of 3.29:
Conclusion:
There is a significant difference in histamine concentration across the four time points.
🔍 Before conducting the hypothesis test, we check:
Independence of observations
➔ !!! ANOVA is NOT robust to violation of this !!!
Normality
➔ ANOVA tends to be relatively robust to non-normality.
Homogeneity of variance (HOV)
➔ ANOVA can handle HOV violations when:
✅ Independence of the Observations:
❓ Then, which assumption should we check for repeated measures ANOVA?
Independency
➔ Do not need to check! (in repeated measures designs)
Normality
➔ The dependent variable (DV) at each level of the independent variable(s) should be approximately normally distributed.
➔ However, ANOVA tends to be relatively robust to non-normality.
Instead of HOV (Homogeneity of Variance)
➔ We should check “Sphericity”
anova_test() function from the rstatix package, which includes Mauchly’s Test of Sphericity.| Dog ID | time1 | time2 | time3 | t1 - t2 | t1 - t3 | t2 - t3 |
|---|---|---|---|---|---|---|
| 1 | 10 | 16 | 25 | -6 | -15 | -9 |
| 2 | 12 | 19 | 27 | -7 | -15 | -8 |
| 3 | 13 | 20 | 28 | -7 | -15 | -8 |
| 4 | 12 | 18 | 25 | -6 | -13 | -7 |
| 5 | 11 | 20 | 26 | -9 | -15 | -6 |
| 6 | 10 | 22 | 27 | -12 | -17 | -5 |
| Variance | 5.367 | 1.6 | 2.167 |
library(tidyverse)
library(plotly)
dat_wide <- tribble(
~ID, ~T1, ~T2, ~T3,
1, 10, 16, 25,
2, 12, 19, 27,
3, 13, 20, 28,
4, 12, 18, 25,
5, 11, 20, 26,
6, 10, 22, 27
)
dat_long <- dat_wide |>
pivot_longer(starts_with("T"), names_to = "Time", values_to = "Y") |>
mutate(Time = factor(Time, levels = paste0("T",1:3)),
ID = factor(ID, levels = 1:6))
dat_long |>
plot_ly(x=~Time, y=~Y) |>
add_markers(color=~ID) |>
add_lines(color=~ID)This table presents:
We are concerned with these variances for “sphericity”
You need your data in long format: one row per subject per time point.
Example:
ANOVA Table (type III tests)
$ANOVA
Effect DFn DFd F p p<.05 ges
1 Time 2 10 221.861 5.2e-09 * 0.95
$`Mauchly's Test for Sphericity`
Effect W p p<.05
1 Time 0.407 0.165
$`Sphericity Corrections`
Effect GGe DF[GG] p[GG] p[GG]<.05 HFe DF[HF] p[HF] p[HF]<.05
1 Time 0.628 1.26, 6.28 2.8e-06 * 0.739 1.48, 7.39 4.28e-07 *This will output:
| Test | Interpretation |
|---|---|
| Mauchly’s Test (p > .05) | Sphericity holds – use standard F-statistic |
| Mauchly’s Test (p < .05) | Sphericity violated – use corrected F-statistics |
If violated, refer to the corrected tests:
epsilon > 0.75Mauchly’s test indicated that the assumption of sphericity had not been violated, W = 0.407, p = .165. We can use standard F-test.
The anova_test() function in the rstatix package does not provide between-subject variability directly in its output. This is because anova_test() focuses on the within-subject effects (repeated measures factors) and treats subject as a blocking factor rather than estimating its variance as a separate effect.
In traditional repeated measures ANOVA, between-subject variability corresponds to:
The variance between the subjects’ overall means — captured by the SS_subjects term.
However, in the anova_test() framework: - The subject-level variability is partialed out (i.e., controlled for), not estimated explicitly. - It is absorbed into the model as the random effect (like a blocking factor in a randomized block design).
You can compute the between-subject sum of squares manually from the subject means using the formula:
SS_{\text{subjects}} = k \sum (\bar{Y}_i - \bar{Y}_{\text{grand}})^2
Where:
# Compute subject means and grand mean
subject_means <- dat_long %>%
group_by(ID) %>%
summarise(subject_mean = mean(Y))
grand_mean <- mean(subject_means$subject_mean)
# Number of repeated measures (e.g., 4 time points)
k <- n_distinct(dat_long$Time)
# Compute SS_subjects
SS_subjects <- subject_means %>%
summarise(SS = k * sum((subject_mean - grand_mean)^2)) %>%
pull(SS)
SS_subjects[1] 20.27778This gives the between-subjects sum of squares.
| Component | Available from anova_test() |
How to Obtain |
|---|---|---|
| Within-subjects | ✅ Effect == "Residuals" |
filter(Effect == "Residuals") |
| Between-subjects | ❌ Not provided | ✅ Compute manually using subject means |